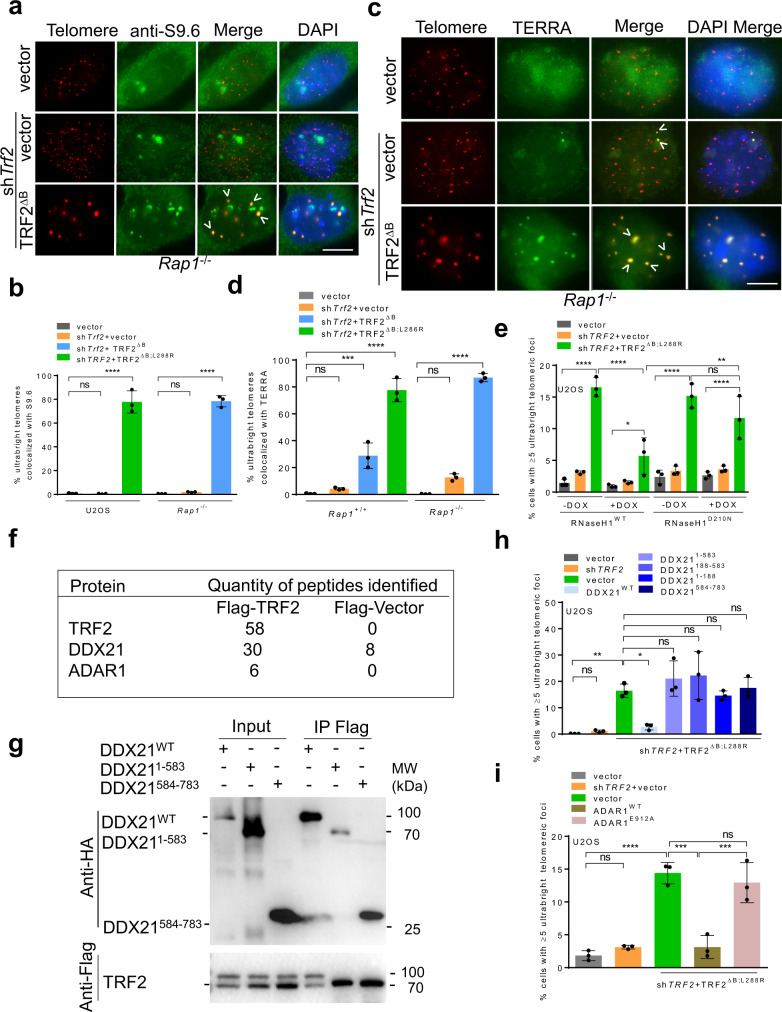
Fig. 5. TRF2B and RAP1 cooperate to repress TERRA and R-loop formation in ultrabright telomeres.
a IF-FISH showing cells containing ≥4 UTs colocalized with S9.6 antibody (green) in Rap1–/– MEFs cells treated with shTrf2 + TRF2ΔB. Scale bars: 5 µm. b Quantification of percent UTs colocalized with S9.6 foci antibody in (a). Data represents the mean of three independent experiments ± SD from a minimum 300 nuclei analyzed per experiment. ****P < 0.0001 by one-way ANOVA. ns non-significant. c IF-FISH showing cells containing ≥4 UTs colocalized with TERRA (green) in Rap1–/– MEFs cells treated with shTrf2 + TRF2ΔB. Scale bars: 5 µm. d Quantification of percent UTs colocalized with TERRA in (c). Data represents the mean of three independent experiments ± SD from a minimum 350 nuclei analyzed per experiment. ***P = 0.0001, ****P < 0.0001 by one-way ANOVA. ns non-significant. e Quantification of UT frequencies after doxycycline induced RNaseH1WT or catalytically dead RNaseH1D210N mutant in U2OS cells expressing indicated constructs. Data represents the mean of three independent experiments ± SD from a minimum of 350 nuclei analyzed per experiment. *P = 0.0325, **P = 0.0035, ****P < 0.0001 by one-way ANOVA. ns: non-significant. f IP-MS data using Flag-TRF2 and Flag-vector as baits showing R-loop associated proteins coprecipitated with Flag-WT TRF2 but not in Flag-vector in one representative experiment. g Co-IP with Flag antibody with lysates from 293 T cells expressing indicated proteins showing the interaction between TRF2 and DDX21 mutant depends on the DDX21 C-terminus. The blot shown is the representative of two independent experiments. h Quantification of UT frequencies in U2OS cells expressing Flag-DDX21WT and the indicated DDX21 deletion mutants in the presence of shTRF2 + TRF2ΔB;L288R. Data represents the mean of three independent experiments ± SD from a minimum 300 nuclei analyzed per experiment. *P = 0.0254, **P = 0.0064 by one-way ANOVA. ns non-significant. i Quantification of UT frequency upon overexpression of GFP ADAR1p110 and catalytically inactive ADAR1p110E912A mutant in U2OS cells expressing indicated DNA constructs. Data represents the mean of three independent experiments ± SD from a minimum 350 nuclei analyzed per experiment. ***P = 0.0001, ****P < 0.0001 by one-way ANOVA. ns non-significant. Source data are provided as a Source Data file.