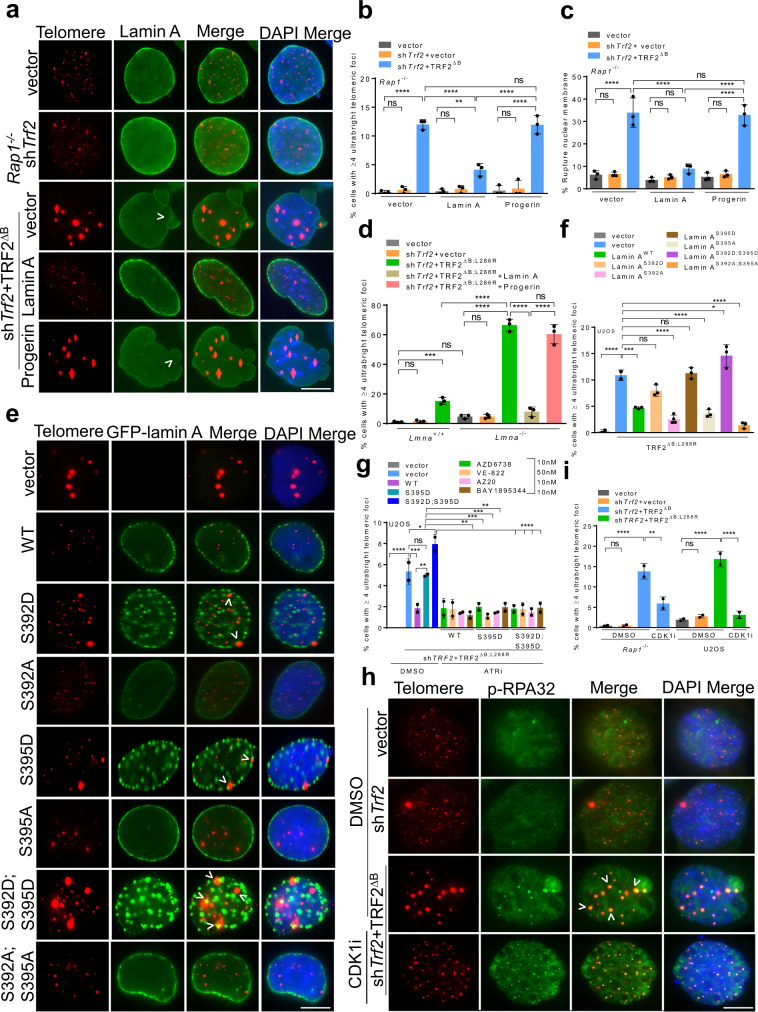
Fig. 7. Defects in lamin A promote the formation of ultrabright telomeres.
a IF-FISH showing lamin A staining (green) in Rap1–/– MEFs expressing indicated constructs and upon expressing WT lamin A or Progerin. Scale bars: 5 µm. b Quantification of UT frequencies in (a). Data represents the mean of three independent experiments ± SD from a minimum 400 nuclei analyzed per experiment. **P = 0.0036, ****P < 0.0001 by one-way ANOVA. ns: non-significant. c Quantification of NE rupturing in cells expressing UTs in Rap1–/– MEFs expressing indicated constructs. Data represents the mean of three independent experiments ± SD from a minimum 400 nuclei analyzed per experiment. ****P < 0.0001 by one-way ANOVA. ns: non-significant. d Quantification of UT frequencies in Lmna+/+ and Lmna–/– MEFs treated with shTrf2 + TRF2ΔB;L286R. Data represents the mean of three independent experiments ± SD from a minimum 300 nuclei analyzed per experiment. ***P = 0.0008, ****P < 0.0001 by one-way ANOVA. ns non-significant. e IF-FISH showing the impact of CDK1 and ATR phospho- and dephospho-GFP-tagged Lamin mutants (green) in the generation of UTs in U2OS cells expressing TRF2∆B;L288R. Scale bars: 5 µm. f Quantification of UT frequencies in (e). Data represents the mean of three independent experiments ± SD from a minimum 350 nuclei analyzed per experiment. *P = 0.0246, ***P = 0.0001 ****P < 0.0001 by one-way ANOVA. ns non-significant. g Quantification of UT frequencies in U2OS cells expressing indicated constructs treated with ATR inhibitors (ATRi). Data represents the mean of two independent experiments ± SD from a minimum 300 nuclei analyzed per experiment. *P = 0.0204, **P = 0.0031, ***P = 0.001, ****P < 0.0001 by one-way ANOVA. ns non-significant. h Representative IF-FISH showing that 4.0 µM Ro-3306 CDK1 specific inhibitor (CDKi) reduced the formation of UTs but not p-RPA32(S33) (green) TIFs in Rap1−/− MEFs expressing indicated proteins. i Quantification of UTs in Rap1–/– MEFs and U2OS cells in the absence and presence of CDK1 inhibitor. Data represents the mean of two independent experiments ± SD from a minimum 300 nuclei analyzed per experiment. **P = 0.0023, ****P < 0.0001 by one-way ANOVA. ns non-significant. Scale bars: 5 µm. Source data are provided as a Source Data file.