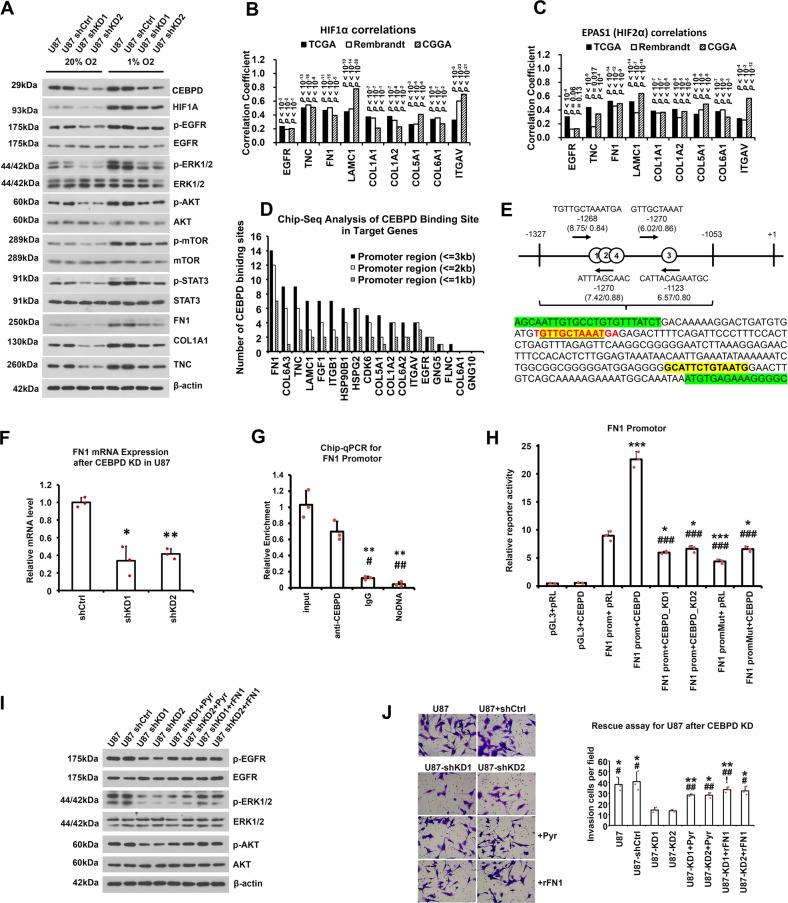
Fig. 6. CEBPD positively regulated ECM-mediated EGFR/PI3K pathway activity, and important genes involved in the pathway and invasion capacity, by directly binding to the promotors of key genes in ECM.
A WB results showing the protein levels of CEBPD and key transducers of the EGFR/PI3K pathway, especially phosphorylated EGFR, ERK1/2, AKT, mTOR, STAT3, and essential genes for the invasion capacity, after CEBPD knockdown in normoxia and hypoxia conditions. B, C Correlation coefficients and corresponding p values (labels above each bar) between HIF1α (B) or EPAS1 (C) and genes in EGFR/PI3K/Akt signal pathways, which are regulated by CEBPD and contribute to tumor invasion capacity. Pearson correlation was performed between HIF1α or EPAS1and indicated genes labeled in the X axis, in three GBM databases (TCGA, Rembrandt, and CGGA databases). D Chip-seq analysis of CEBPD binding sites in CEBPD-regulated proteins belong to the EGFR/PI3K pathway. Bars represented number of CEBPD binding sites identified by Chip-seq in the promoters of the indicated proteins. E The DNA fragment of CEBPD binding peaks in the FN1 promoter identified by Chip-seq, which also contained highly reliable CEBPD binding sites as predicted by the JASPAR database [43]. There are 4 highly reliable CEBPD binding consensus sequences (red letters, site 1–4) in the fragment, and the site (1) and (2) had the greatest scores. For each binding site, the sequence, location, and the binding scores (expressed as score/relative score) were labeled; red letters represent positive strand, and yellow shaded letters represent negative strand. Greed shaded letters represent sequences for primers. F mRNA levels of FN1 after shCEBPD treatment in U87 cells in normoxia condition. G Chip-qPCR showing that CEBPD bind to the FN1 promoter region shown in E in normoxic condition. H Luciferase reporter assay showing bind and activation of CEBPD on the promotor of FN1 in normoxia condition. The promoter region of FN1 is shown in (E). For mutation analysis, the putative binding sites (site 1 and 2) with the greatest scores were mutated, where the sequence of “TGTTGCTAAATGA” (as shown in E) were replaced by “CTGCCGCGGCGGG”. (pGL3: pGL3-Basic vector containing blank control; FN1 prom: pGL3-Basic vector containing FN1 promoter sequence; FN1 promMut: pGL3-Basic vector containing FN1 promoter with mutation; pRL: pRL-TK vector containing blank control; CEBPD: pRL-TK vector containing CEBPD sequence; CEBPD_KD: pRL-TK vector containing CEBPD shRNA sequence.) *p < 0.05, ***p < 0.0001 compared to the FN1 promoter + pRL group; ###p < 0.0001 compared to the FN1 promoter + CEBPD group. All of the last 6 groups had p < 0.0001 compared to the first 2 groups. I WB assays of rescue experiments after CEBPD KD in U87 cells. Protein levels of key transducers of the EGFR/PI3K pathway, including p-EGFR, were examined in U87 cells, U87 cells treated with shCtrl (U87 shCtrl) and shCEBPD lentivirus (U87 shKD1, U87 shKD2), or U87-shCEBPD KD cells which are recused with the α5β1 integrin agonist Pyrintegrin (U87 shKD1+Pyr, U87 shKD2+Pyr) or a recombinated FN1 protein (U87 shKD1 + rFN1, U87 shKD2 + rFN1). The α5β1 integrin is the receptor for FN1. J Transwell invasion assays of rescue experiments after CEBPD KD for U87 cells. The cell groups were the same as described in (I), and experiments were repeated in triplicate.