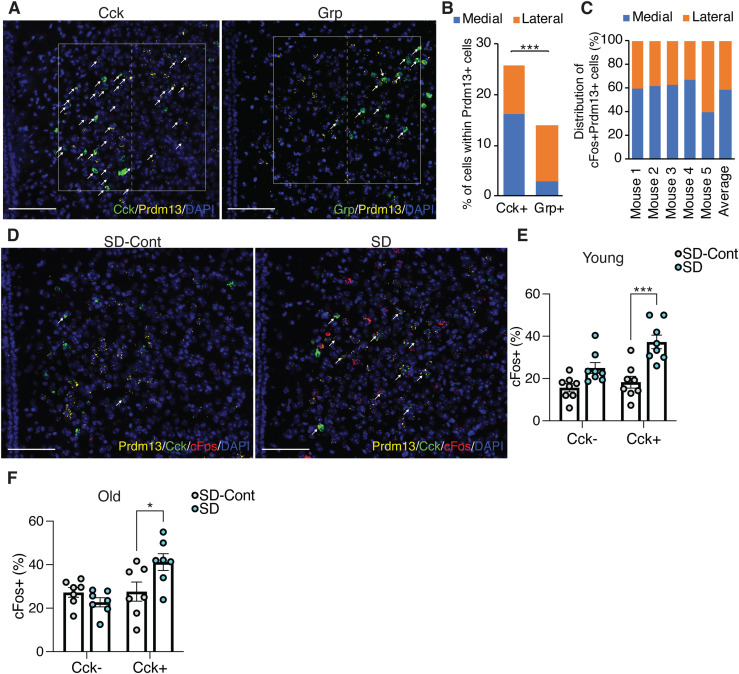
Figure 8. DMH-Prdm13+Cck+ neurons are activated during SD.
(A) Representative images of the DMH with Prdm13 (yellow) and one of the two genes, Cck or Grp (green) visualized by RNAscope. Cells were counterstained with DAPI (blue). White boxes show the DMH, which is divided into medial and lateral areas by dashed lines. White arrows show yellow+green+ cells. Scale bar indicates 100 μm. (B) Ratios of Cck+ or Grp+ cells within Prdm13+ cells in medial, lateral, or total (medial and lateral) DMH (n = 3–5). Values are shown as means ± S.E., ***P < 0.001 by unpaired t test. (C) Distribution of cFos+Prdm13+ cells (n = 5). (D) Representative images of the DMH from young mice under SD-Cont and SD with Prdm13 (yellow), Cck (green), and cFos (red) visualized by RNAscope. Cells were counterstained with DAPI (blue). White arrows show Prdm13+Cck+cFos+ cells. Scale bar indicates 100 μm. (E, F) Ratios of cFos+ cells within Prdm13+Cck− (left) or Prdm13+Cck+ (right) cells in young (E) or old (F) mice during SD-Cont and SD (n = 7–8). Values are shown as means ± S.E., *P < 0.05, ***P < 0.001 by two-way ANOVA with Bonferroni’s post hoc test.