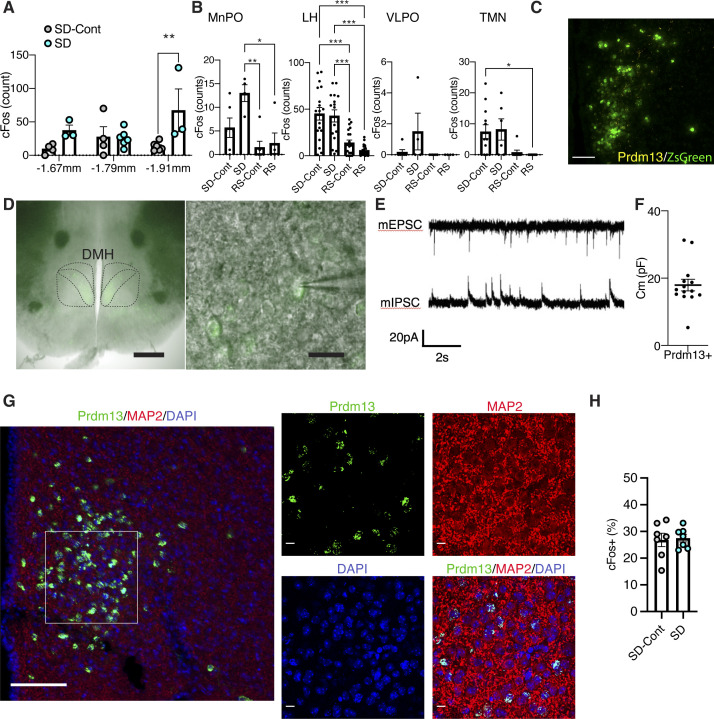
Figure S2. Neurons are activated in the DMH during SD, and Prdm13+ cells are electrically active.
(A) Numbers of cFos+ cells in the DMH at bregma −1.67, −1.79, and −1.91 mm during SD and sleeping control (SD-Cont) detected by cFos immunohistochemistry (n = 3–6). Values are shown as means ± S.E., **P < 0.01 by two-way ANOVA with Bonferroni’s post hoc test. (B) Numbers of cFos+ cells in the median preoptic area (MnPO), lateral hypothalamus (LH), ventromedial preoptic area (VLPO), and tuberomammillary nucleus during SD, recovery sleep (RS), and sleeping control (SD-Cont and RS-Cont) detected by cFos immunohistochemistry (n = 4–22). Values are shown as means ± S.E., *P < 0.05, **P < 0.01, and ***P < 0.001 by one-way ANOVA with Bonferroni’s post hoc test. (C) Representative image of the DMH with ZsGreen fluorescence derived from Prdm13-CreERT2 (green) and mRNA of Prdm13 (yellow) visualized by RNAscope. Scale bar indicates 100 μm. (D) Representative images showing Prdm13+ neurons in the DMH (left). High-magnification images of Prdm13+ DMH cells (right). Scale bar indicates 500 and 20 μm (left and right, respectively). (E) Representative mEPSC and mIPSC traces obtained from Prdm13+ DMH cells. (F) Membrane capacitance of Prdm13+ DMH cells recorded in whole-cell patch (n = 14). Values are shown as means ± S.E. (G) Representative image of the DMH with ZsGreen fluorescence derived from Prdm13-CreERT2 (green) and neuronal marker MAP2 (red). Cell nuclei were counterstained by DAPI (blue). Boxed areas were shown at high magnification. Scale bar indicates 100 (low magnification) and 10 μm (high magnification). (H) Ratios of cFos+ cells within Prdm13+ cells in old mice during SD and SD-Cont detected by RNAscope in situ hybridization (n = 7). Values are shown as means ± S.E.