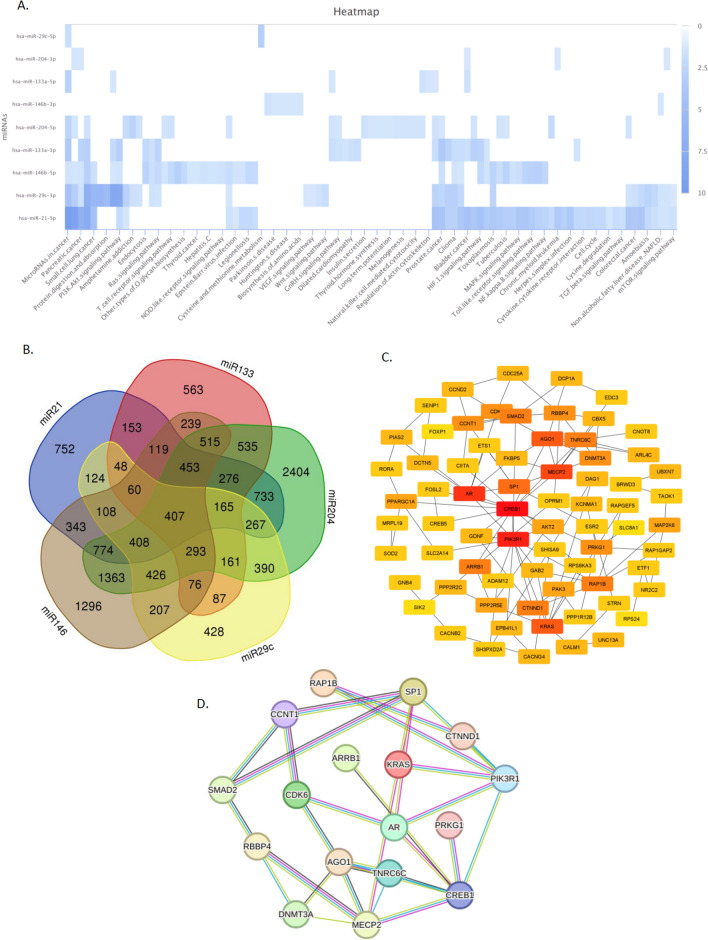
Figure 5.
(A) Heat map of highly validated miRNAs and pathway they involved using miRPathDB v2.0 (https://mpd.bioinf.uni-sb.de/heatmap_calculator.html?organism=hsa) (B) candidate miRNAs and their common target genes using Venn diagram online database (https://bioinformatics.psb.ugent.be/webtools/Venn/); (C) 100 highly scored genes based on degree were selected using Cytohubba tools (https://cytoscape.org/ cytoscape version3.9.1) (D) miRNA-target genes; Hub genes reanalyzed by string database consist of 100 nodes and 223 edges. We set the highest confidence score of 0.9 and hide disconnected nodes in the network (https://string-db.org/).