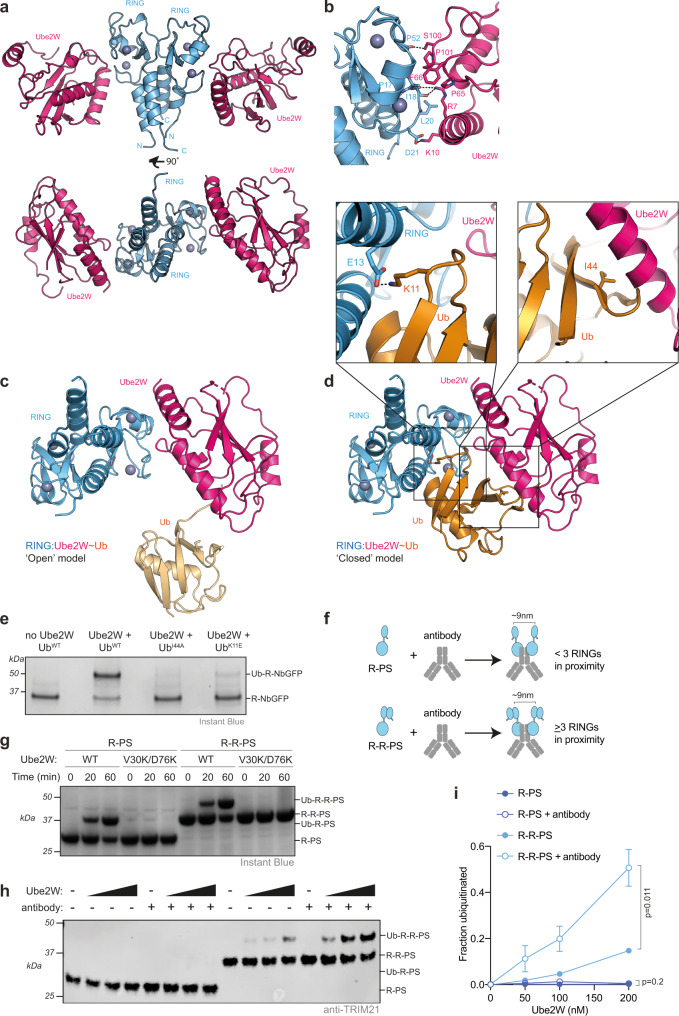
Fig. 1. Dimeric Ube2W and RING clustering is required for ligase autoubiquitination.
a 2.25 Å X-ray structure of TRIM21 RING (blue) in complex with Ube2W (pink). b Close-up of the E2:E3 interface. c, d Structural models of a RING:Ube2W~Ub complex with Ub in an open (c) or closed (d) conformation respectively. Based on superposition with the Ub-RING:Ube2N~Ub:Ube2V2 structure 7BBD39. In the closed conformation (d; boxed), Ub K11 makes a potential salt bridge with TRIM21 E13 and buries I44 at an interface with the E2. e Ube2W-mediated TRIM21 RING mono-ubiquitination assay using 2 µM R-NbGFP and 1 µM Ube2W with WT, I44A or K11E ubiquitin. Representative example from n = 2 independent experiments. f Schematic of antibody-induced recruitment of either two RINGs or two RING dimers. Only the latter satisfies the ‘two-plus-one’ model for RING autoubiquitination20,39. g Ube2W-mediated mono-ubiquitination assay using 10 µM R-PS or R-R-PS and 0.25 µM Ube2W WT or monomeric V30K/D67K. Representative example from n = 2 independent experiments. h Ubiquitination of 100 nM R-PS or R-R-PS in the absence or presence of equimolar anti-GFP antibody. Ube2W was titrated (25, 50, 100, 200 nM). Representative example from n = 3 independent experiments. i Quantification of monoubiquitination from (h). Graph shows mean and s.e.m. from n = 3 independent experiments. Statistical significance based on two-tailed Student’s t test (two-tailed). Source data are provided as a Source Data file.