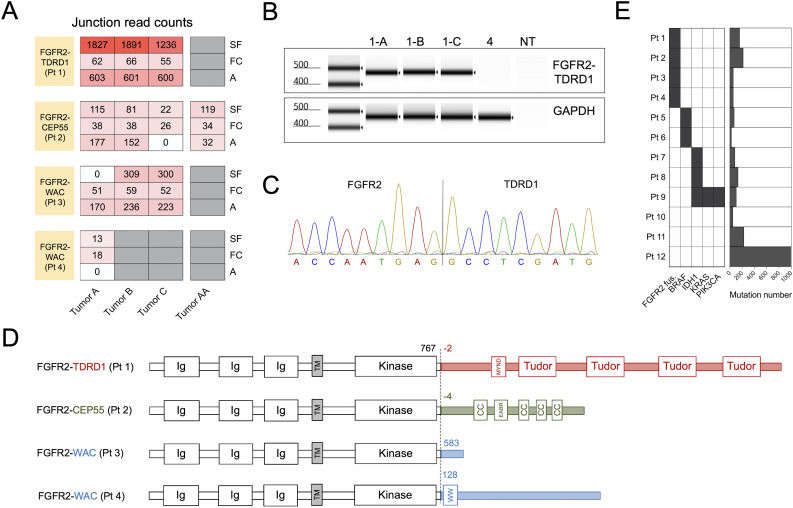
Figure 1.
Tumors from four patients with metastatic intrahepatic cholangiocarcinoma (ICC) harbored in-frame FGFR2 gene fusions. (A) Patients’ metastases were surgically resected and subjected to whole transcriptome sequencing. Three different computational algorithms (‘A’, Arriba; ‘FC’, FusionCatcher; and ‘SF’, STAR-Fusion) were used to identify in-frame gene fusions involving the FGFR2. The heatmap depicts junction read counts (ie, reads spanning fusion breakpoints) reported by each fusion caller (rows) for all the tumor samples from four FGFR2 fusion-positive patients (columns). For each patient, all the tumors were resected at the same time, except for patient 2, who had a resection prior to the administration of adoptive T cell therapy targeting ERBBI2PE805G mutation (Tumor AA), and a repeat resection after the therapy (Tumors A–C). (B) RT-PCR was performed on tumor cDNA using primers that flanked the breakpoints of each FGFR2 fusion. Result of automated electrophoresis of the FGFR2-TDRD1 amplification reaction from patient 1’s tumors (1-A, 1-B and 1-C) is depicted. cDNA from patient 4’s tumor was used as a negative control, along with the reaction with no added template (NT). Primers for the wild type human GAPDH were used to generate a positive control. (C) Amplified products from (B) were purified and subjected to Sanger sequencing. The chromatogram depicts the sequencing result for tumor A from patient 1 (same sequence was detected in tumors B and C). (D) Breakpoint coordinates of each FGFR2 fusion transcript were used to reconstruct the sequences of FGFR2 fusion proteins. The dashed line indicates the fusion breakpoints; the numbers next to it indicate the amino acid position at which wild type FGFR2 protein terminates (black) or at which the partner protein starts (colored). The names of the protein domains are provided in the boxes. CC, coiled-coil domain; Ig, immunoglobulin-like domain; MYND (myeloid, Nervy, and DEAF-1)-type zinc finger domain; TM, transmembrane domain. Note that FGFR2 and partner proteins are not depicted on the same scale for clarity purposes. (E) The left panel depicts distribution of FGFR2 fusions and driver (hotspot) point mutations in the study cohort. The hotspot mutations include BRAFD594N (patient 5), BRAFV600E (patient 6), IDH1R132C (patients 7–9), and KRASG12V and PIK3CAH1047R (patient 9). The right panel depicts the overall number of tumor-specific mutations (including single nucleotide variants and insertions/deletions) in the same cohort. cDNA, complementary DNA; FGFR2, fibroblast growth factor receptor 2; TDRD1, tudor domain-containing 1.