Abstract
Autophagy is a cellular homeostasis mechanism that fuels the proliferation and survival of advanced cancers by degrading and recycling organelles and proteins. Preclinical studies have identified that within an established tumor, tumor cell autophagy and host cell autophagy conspire to support tumor growth. A growing body of evidence suggests that autophagy inhibition can augment the efficacy of chemotherapy, targeted therapy, or immunotherapy to enhance tumor shrinkage. First-generation autophagy inhibition trials in cancer using the lysosomal inhibitor hydroxychloroquine (HCQ) have produced mixed results but have guided the way for the development of more potent and specific autophagy inhibitors in clinical trials. In this review, we will discuss the role of autophagy in cancer, newly discovered molecular mechanisms of the autophagy pathway, the effects of autophagy modulation in cancer and host cells, and novel autophagy inhibitors that are entering clinical trials .
Keywords: Autophagy, Cancer, Immunotherapy, Chemotherapy, Hydroxychloroquine, Lysosome
Autophagy as a potential therapeutic target in cancer
The abnormal growth of cancer cells requires more nutrients than normal cells. Therefore, the pathways which control cellular growth and metabolism are a major focus of therapeutic drug development. Autophagy is an evolutionarily conserved intracellular degradation mechanism that recycles intracellular components to maintain homeostasis and to promote cell survival during stress [1]. The recycling capacity of autophagy arguably makes it a growth pathway and provides an internal source of nutrients to the growing cancer cell. In the harsh conditions of the tumor microenvironment (TME, see Glossary), which has disordered vasculature and hypoxia, this internal source of nutrients can play a critical role for tumor cell survival [2,3]. Autophagy was originally referred to as a tumor-suppressor mechanism, that prevents the accumulation of damaged organelles and subsequent genomic instability which is critical for tumorigenesis [4-6]. However, more recent evidence from mouse models detailed below suggests that autophagy does not play a strong tumor suppressor role in cells lacking multiple oncogenic mutations [7,8]. In fact, in the setting of advanced cancer, autophagy does not impair but promotes tumor cell survival [9-12]. Understanding the role of autophagy in different stages of tumorigenesis is important when trying to decide as a field whether to develop autophagy inhibitors or inducers as pharmacological agents to target autophagy in cancer.
Genetically engineered mouse models of cancer are the setting in which this controversy of whether autophagy should be promoted or inhibited in cancer came to light. For example, in a mouse model of pancreatic cancer (Kras mutant Tp53−/−) with defective autophagy-related (Atg) gene 5 (Atg5) deleted, mice did not die of pancreatic cancer, but rather died of pancreatic polyps (benign tumors) overwhelming the pancreas [13]. In human cancer, very few cells destined to become tumors carry oncogenic mutations and non-cancerous cells do not; but in Kras mutant Tp53−/− Atg5-null model all pancreatic cells had oncogenic mutations questioning the relevance of this model to human cancer. When autophagy deficiency was modeled in pancreatic cells without mutations in Kras or Tp53, no benign tumor growth was initiated [14]. This finding put to rest the concern that effective drugs that inhibit autophagy will induce tumorigenesis in patients. On the other hand, genetic or pharmacological autophagy inhibition has been shown to slow down tumor growth and prolong overall survival in numerous mouse models of advanced cancer [14-18].
Another major advance in understanding the role of autophagy in cancer is that autophagy in the non-mutated host cells in the TME also supports tumor growth [19-23]. A pharmacological autophagy inhibitor or inducer would modulate autophagy in all cells in the body, not just the tumor cells. Since autophagy inhibition in normal cells does not induce tumorigenesis, and autophagy inhibition in cancer cells promotes survival [14], these findings provide further rationale for focusing on autophagy inhibitors and not autophagy inducers for advanced cancer. Patients with advanced cancer often are willing to tolerate high rates of side effects in return for shrinkage of the tumor, but if targeting autophagy produces catastrophic toxicity, there is no chance for effective drug development. In a mouse model of lung cancer, in which autophagy was completely inhibited throughout the body, mice could tolerate a complete lack of autophagy for 2-3 months but fatal multisystem organ failure ensued eventually in these mice [15]. Although drug therapy can never achieve complete autophagy inhibition like genetic deletion can, clinical development of autophagy-modulating therapy needs to be done carefully.
Initial efforts in translating proof-of-principle in vivo studies, demonstrating that blocking autophagy enhances cancer therapy into clinical trials, focused on the lysosomotropic drug hydroxychloroquine (HCQ). The initial phase I trials of HCQ in combination with standard-of-care treatments demonstrated that autophagy inhibition can be achieved in patients without excess toxicity [24]. In some but not all clinical trials there is some evidence that the addition of HCQ to standard-of-care therapy can produce clinical antitumor activity [25-28]. However, both in pre-clinical models and in clinical trials, autophagy inhibition with HCQ alone or in combination is not curative, suggesting there are resistance mechanisms to this approach [24,25,28-30].
Even though there is a large amount of evidence that autophagy is an important pathway in cancer, neurodegeneration, and infectious disease [1], to date no specific autophagy inhibitor or inducer has received regulatory approval in cancer or any other disease. These issues raise the need to find better pharmaceuticals for targeting autophagy in cancer. In this review, we will highlight newly discovered regulatory mechanisms of mammalian autophagy that could in the future serve as targets for drug discovery, different contexts in cancer where autophagy modulation may be safe and effective, and therapeutics that are emerging in preclinical studies and clinical trials to modulate autophagy in cancer.
Newly elucidated regulatory mechanisms of autophagy
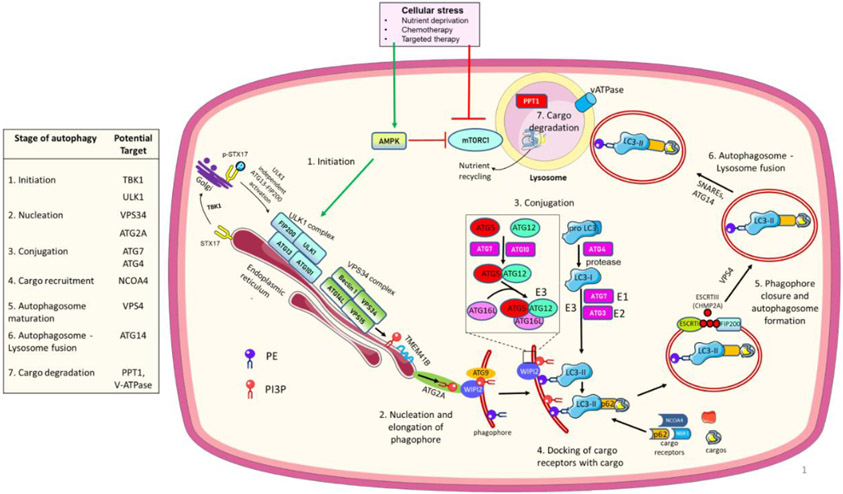
The autophagy pathway has been studied in yeast and in eukaryotic cells for decades, but especially in mammalian cells, new regulatory elements are being discovered that may have implications for drug development. To understand the significance of the new discoveries we first will highlight a basic understanding of the canonical autophagy pathway, which consists of the creation of a circular doublemembrane vesicle called an autophagosome that sequesters and delivers cytoplasmic cargo for degradation in the lysosome (Figure 1).
Figure 1. Autophagy regulatory pathway.
Cellular stress activates AMPK which directly and indirectly (through inhibition of mTORC1) activates the ULK1 complex and initiates autophagy. An ULK1 independent activation of autophagy can also occur via TBK1. ULK1 phosphorylates and activates the PI3KC3 lipid kinase complex which generates phosphatidylinositol 3-phosphate (PI3P) to commence phagophore membrane elongation with the help of TMEM41B and ATG2A, which deliver lipids to PI3P-WIPI2 complexes for the next conjugation steps. Two ubiquitin-like sequential conjugation reactions (ATG12 and ATG7 conjugate ATG8 (LC3) protein to phosphatidylethanolamine (PE). ATG7 and ATG3 act as E1 and E2-like enzymes, respectively, and the ATG12-ATG5-ATG16L complex as an E3-like enzyme to finally catalyze the binding of LC3I to PE by ATG3. Next, autophagy cargo receptors with their specific cargos dock onto LC3II in the phagophore/growing autophagosome. ESCRT-III protein polymerization brings together the open ends of the autophagosome which are closed by VPS4 to complete the autophagosome maturation. The autophagosome fuses with the lysosome to degrade and recycle autophagy cargo components. Proteins in the table present potential targets to inhibit different autophagy steps in cancer. For recent advances in each step, please see the main text.
The initiation of autophagy begins when either the nutrient sensor mammalian target of rapamycin complex 1 (mTORC1) is inhibited or the energy sensor AMP-activated protein kinase (AMPK) is activated. Metabolic or therapeutic stress that leads to either activation of AMPK or inhibition of mTORC1 results in an activated Unc51-like kinase 1 (ULK1) complex, the first complex in the autophagy pathway. When the ULK1 complex is activated, it phosphorylates and activates the vacuolar protein sorting 34 (VPS34) complex. The VPS34 complex has lipid kinase activity that prepares membranes for assembly of the ATG8/light chain 3 (LC3) conjugation machinery, which includes drug targets ATG4 and ATG7. This conjugation machinery acts like a ubiquitin ligase system but uniquely conjugates LC3 family members (such as LC3B) to lipid constituents on the forming autophagosome. Once LC3B is attached to the autophagosome membrane it not only serves as a marker of autophagy but a docking site for autophagy cargo receptors SQSTM1 (p62), NCOA4, and the neighbor of BRCA1 gene (NBR1) that sequester cargos like organelles and proteins that are marked by ubiquitination for their autophagic degradation. Autophagosome maturation is followed by fusion with the lysosome. The lysosome is an acidic organelle because of the presence of a proton pump, the vacuolar -ATPase (V-ATPase). Once an autophagosome fuses with the lysosome, acid-dependent enzymes such as cathepsins degrade autophagic cargo and nutrient transporters recycle macromolecules that fuel growth of the cell. For a comprehensive review of the molecular machinery of autophagy please see [1].
Although the basic mechanism of autophagy has been known for years, a number of recent studies related to each step provide potentially new autophagy regulators as targets for drug development (Figure 1). Related to autophagy initiation (Step 1), a recent study demonstrated that tank-binding kinase 1 (TBK1) phosphorylates Syntaxin17, which can then assemble an ATG13-FIP200 protein complex independent of ULK1 activity to activate autophagy [31]. A novel regulator of autophagosome biogenesis (Step 2) is the endoplasmic reticulum resident transmembrane protein 41B (TMEM41B), which mobilizes lipids into the emerging autophagosomal membrane [32,33]. The elongation of emerging autophagosome (Step 2) is regulated by recently identified interaction between ATG2A and a lipid scramblase, ATG9. ATG2A delivers lipids to PI3P-WD repeat domain phosphoinositide-interacting (WIPI2) complexes, and ATG9 rearranges the lipids into a symmetrical bilayer that feeds into the autophagosome [34]. Maturation of the autophagosome (Step 5) is carried out by the recruitment of endosomal sorting complexes required for transport (ESCRT) proteins. ESCRT-1 recruits the filament-forming charged multivesicular body protein 2A (CHMP2A), with the help of FIP200. Filament polymerization brings together the open ends of the autophagosome followed by membrane fission. An AAA-ATPase VPS4 finally settles the fission process and closes the autophagosome [35]. Previously, SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) [36,37] were thought to be the key drivers of autophagosome-lysosome fusion (Step 6). However, a recent paper identified ATG14 as a bifunctional autophagy protein that was not only required for the initiating ULK1 complex but also for efficient autophagosome-lysosome fusion [38].
Finally, a recent study identified Lysosomal Enzyme Trafficking Factor (LYSET), a transmembrane protein in the Golgi that is required for lysosomal enzymes trafficking to the lysosome to enable lysosomal degradation (Step 7) [39]. One such lysosomal enzyme is palmitoyl-protein thioesterase 1 (PPT1), which was found to regulate lysosomal acidification by controlling the localization of palmitoylated subunits of the V-ATPase [40]. In addition, mTORC1 was recently found to regulate the assembly of V-ATPase [41]. Although not many new discoveries have been about the core autophagy conjugation machinery (Step 3-4), there is an emerging literature that there are autophagy-independent roles of certain ATG genes such as ATG5 and ATG7 including LC3-associated phagocytosis [42], and these roles may be very important in cancer progression [43]. Multiple autophagy genes, not just ATG7 or ATG5, need to be tested in mechanism-focused laboratory research that is attempting to implicate autophagy in a biological process. Specifically, genetic or chemical modulation of one protein in the ULK1 complex, the VPS34 complex, the LC3 conjugation cascade, and the lysosome would be the most comprehensive way to imply that autophagy has a role in any given biological process.
Targeting tumor cell autophagy in combination with chemotherapy or targeted therapy
Patients with advanced cancer are treated with chemotherapy, targeted therapy, or immunotherapy, but in most cases, these treatments are not curative. Autophagy promotes the survival and resistance of cancer cells in the face of chemotherapy and targeted therapy. Its role in impairing anti-tumor immunity is only recently being appreciated. In each therapeutic context autophagy inhibition has been shown to enhance the efficacy of the anti-cancer modality mostly in preclinical models, but also in some clinical trials.
Autophagy inhibition augments chemotherapy
Multiple in vitro and in vivo studies suggest that traditional cytotoxic chemotherapy drugs induce autophagy in multiple cancer types and blocking autophagy with the lysosomal inhibitors chloroquine (CQ) or HCQ can augment chemotherapy efficacy [44]. The most compelling evidence that this can be translated into patient benefit comes in pancreatic ductal adenocarcinoma (PDAC). In a phase I/II trial of neoadjuvant HCQ and gemcitabine (i.e., given prior to surgery) was tolerable and produced a 61% reduction in the pancreatic cancer biomarker CA19-9 (cancer antigen 19-9) at the time of surgery [45]. This was followed by a randomized phase II clinical trial of neoadjuvant gemcitabine and nab-paclitaxel therapy, with or without HCQ in resectable PDAC [26]. Patients received standard adjuvant therapy but not HCQ after surgery. In this larger clinical trial, the HCQ + chemotherapy arm produced a significantly higher pathological response rate compared to the chemotherapy alone arm. However, there was no significant difference in relapse-free and overall survival indicating a possible development of drug resistance, or the mitigation of HCQ effects with time. A recent clinical update found that 31% of locally advanced PDAC patients treated with HCQ + gemcitabine survived > 5 years [25]. A correlative study looking at both of these neoadjuvant trials found that patients with SMAD4 transcription factor deficiency in their tumors had a higher rate of benefit with an HCQ based regimen compared to patients who did not have a SMAD4 deficient tumor [46]. Finally, a randomized phase II trial in Stage IV pancreatic cancer comparing gemcitabine/nab-paclitaxel + HCQ to gemcitabine/nab-paclitaxel also found a higher response rate but no difference in progression-free or overall survival by the addition of HCQ [28]. Taken together, these findings support that autophagy inhibition with chemotherapy could provide benefits for PDAC treatment, but perhaps a more potent and specific autophagy inhibitor is needed to produce superior durability of response compared to HCQ.
Autophagy inhibition augments targeted therapy
The most striking autophagy activation is seen in the setting of mitogen-activated protein kinases (MAPK) pathway inhibition [47]. MAPK signal transduction cascade progresses via sequential phosphorylation of RAS, RAF, MEK and ERK kinases. The first node of MAPK signaling link to autophagy was BRAF (v-raf murine sarcoma viral oncogene homolog B1), a serine/threonine protein kinase that plays a critical role in the MAPK signaling [48].
Although FDA-approved BRAFV600 inhibitors (BRAFi) combined with MEK inhibitors (MEKi) have revolutionized the treatment strategy of BRAF-mutated cancers, primary and acquired chemoresistance remains a major obstacle for patients [49]. Autophagy has been reported as a major druggable resistance mechanism to targeted therapy or in BRAF-mutant cancers by many groups [12,50-52]. This has led to the launch of the non-randomized BAMM (BRAF, Autophagy and MEK Inhibition in Melanoma) phase I/II clinical trial of dabrafenib (BRAFi), trametinib (MEKi) with HCQ [53]. The triple drug combination was tolerable and produced an 85% response rate in stage IV BRAF mutant melanoma. A high response rate was also observed in patients with elevated serum lactate dehydrogenase (LDH), which is an extremely poor prognostic marker in melanoma. Considering the safety and response rate achieved in BAMM trial, a double-blind randomized trial (BAMM2/EA6191) combining dabrafenib and trametinib with HCQ or placebo has been launched (NCT04527549) to discern whether the addition of HCQ to BRAF and MEKi is truly beneficial.
The other major MAPK genotype in which autophagy plays a role is RAS mutant cancers [47]. Several reports suggested that autophagy inhibition can synergize with the MEK or ERK inhibitors in RAS mutant cancers. A genetic screen demonstrated that combined knockdown of autophagy and MAPK components produced the maximum cytotoxicity in RAS mutant cells [54]. Kinsey et al. found that the MEKi trametinib induced autophagy in RAS mutant cancer lines. The combination of CQ and trametinib produced synergistic antitumor efficacy in RAS mutant PDX tumors [55]. Similarly, ERK inhibition in KRAS-driven human PDAC cells induces autophagy and targeting ERK with CQ produced additive anti-tumor activity in vivo [56]. Ongoing clinical trials combining MEKi or ERKi with HCQ are evaluating the safety and efficacy of this approach (e.g., NCT04132505, NCT04386057).
The promising preclinical and clinical data from BRAF and RAS mutant cancer studies combining MAPK pathway and autophagy inhibitors suggests that this approach may produce benefits. However, a mechanistic connection between autophagy and MAPK pathway is still being investigated and may allow the development of more specific and efficient inhibitors for treating such cancers. In a recent study, a decrease in MYC following MAPK inhibition in PDAC cells enabled unrestricted access of MiT/TFE (the microphthalmia/transcription factor E) transcription factors to CLEAR (Coordinated Lysosomal Expression and Regulation) network transcriptional elements with a corresponding elevation in autophagy and lysosomal gene expression [57]. Two different studies showed that MEKi in PDAC cells upregulates ferritinophagy, an autophagy process that selectively degrades the iron storage protein ferritin in the lysosome to release iron in the cytosol [57-59]. This increase in iron promotes mitochondrial iron-sulfur cluster protein synthesis for enhanced mitochondrial oxidative phosphorylation in PDAC cells [57-59]. Targeting ferritinophagy could be an interesting new approach to autophagy inhibition but requires more research.
Autophagy inhibition in the TME augments anti-tumor immune responses
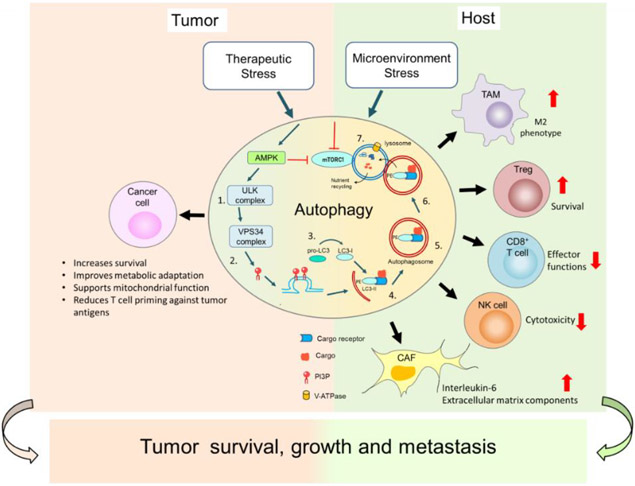
Immunotherapeutic strategies aim to boost anti-tumor immunity in cancer; however, the clinical outcomes have been less effective than anticipated. Recent findings suggest autophagy inhibition not only in tumor cells but in different cells present in the TME such as T cells, and tumor-associated macrophages (TAMs) could augment anti-tumor immunity and enhance immune checkpoint inhibitors therapies, such as antiprogrammed cell death protein 1 (PD-1) antibody treatment [60,61]. These findings emphasize that autophagy in both the tumor cells and host cells supports tumor growth (Figure 2).
Figure 2. Autophagy supports tumor growth and survival by modulating cancer cells in the tumor, and host cells in the tumor microenvironment (TME).
Stressors on cancer cells induce autophagy (please see Figure 1 for details) in both cancer cells as well as in non-cancerous host cells. In cancer cells, autophagy induction promotes their survival by altering their metabolism to provide nutrients and promoting their mitochondrial function under stress. Autophagy degrades immunoproteasome components and MHC-I molecules to escape antitumor immunity. In the TME in non-cancerous host cells, autophagy promotes an M2 phenotype of tumor associated macrophages (TAMs) that support tumor cell growth. Autophagy can also enhance T regulatory cells (Tregs) leading to immunosuppression in the TME. Autophagy limits infiltration and effector functions of CD8+ T cells and natural killer (NK) cells. Autophagy also enhances the activity of cancer associated fibroblasts (CAFs) in the extracellular matrix that supports tumor growth and metastasis. Together the functions of autophagy in cancer cells and host cells support tumor survival, growth, and metastasis.
Autophagy inhibition impairs T regulatory cells (Tregs) survival and reduces cytotoxic T-cell exhaustion
Studies show that the extent of somatic DNA mutations in cancer cells dictates the mechanism of anti-tumor activity shown by host cells lacking autophagy, either enhancing antitumor immunity [62] or by reducing an oncometabolite production [63]. In one study when host animals with a whole-body deletion of Atg7 gene were implanted with liver cancer cells containing high tumor mutational burden (TMB); and therefore, a larger number of neoantigens for the immune system to recognize, a T cell-dependent anti-tumor immune response ensued that was characterized by reduced Tregs and intra-tumoral T-cell exhaustion [62]. Similar to these findings, in a breast cancer model, autophagy was found to impair anti-tumor effector functions of CD8+ T cells. CD8+ T cells lacking Atg5 gene were more metabolically active, produced higher levels of interferon-γ (IFN-γ) and tumor necrosis factor α (TNF-α) and displayed an effector antitumor memory phenotype [64].
It is still unclear if autophagy inhibition only augments anti-tumor immunity in high TMB cancers. For instance, in one study, animals harboring low TMB liver cancer cells did not show an increased immune response with whole body Atg7 knockout [63]. Tumors did shrink with Atg7 deletion, but it was due to a metabolic compromise to the tumor cells. Loss of autophagy in normal hepatocytes resulted in reduced circulating levels of arginine which is an essential amino acid for tumor cells growth [63]. In contrast, the ability of autophagy to suppress an anti-tumor immune response was also uncovered by a genome wide CRISPR screening in commonly used mouse cancer models that are typically low TMB compared to human cells [65]. Taken together these findings suggest more work needs to be done in human tissues to understand the role of autophagy in regulating T cell immunity against cancer cells in high versus low TMB tumors.
Autophagy inhibition enhances antigen presentation by tumor cells
An adaptive immune response to cancer cells depends on antigenic peptide epitopes generated and presented by cancer cells via major histocompatibility complex (MHC) molecules to CD4+ and CD8+ T cells. Two recent studies have implicated autophagy in regulating antigen presentation.
Deng et al. demonstrated that autophagy specifically degrades components of the immunoproteasome in the cancer cell, which generate peptides presented by MHC. Inhibition of ULK1 blocked autophagic degradation of immunoproteasome components and enhanced the efficacy of PD-1 antibody in a liver kinase B1 (LKB1) mutant nonsmall cell lung cancer mouse model. This genotype is particularly immunotherapy resistant in humans and is especially sensitive to genetic autophagy inhibition [66].
One of the most potentially impactful ways that autophagy regulates tumor immunity is through its regulation of MHC class I. Yamamoto et al. demonstrated that MHC-I molecules are selectively targeted for lysosomal degradation by an autophagy-dependent mechanism that involves the autophagy cargo receptor NBR1 [67]. Blocking autophagy genetically or with CQ increased surface expression of MHC Class I improving T cell priming. Combining CQ with anti-PD-1 and anti-CTLA4 antibodies produced synergistic anti-tumor activity in an immune-cold mouse model of pancreas cancer [67]. Taken together, there appear to be multiple ways that autophagy can regulate antigen presentation, but confirmation in human tissues and in other cancer types needs to be done and is an area of active research.
Autophagy in TAMs
TAMs are a vital component of the TME, and sometimes make up to 40-50% of the tumor mass in certain tumors such as melanoma [68]. Depending on the phenotype, M1 or M2, TAMs display anti-tumor or pro-tumorigenic properties, respectively. Autophagy inhibition in TAMs promotes M1 polarization, resulting in increased proinflammatory cytokine secretion [20]. In melanoma models, HCQ treatment repolarizes macrophages from an M2 to M1 phenotype while also upregulating the stimulator of interferon gene (STING). Upregulated STING results in the phosphorylation of TBK1 and release of interferon β. TAM-induced interferon secretion enhances tumor-directed cytotoxic T cells, and HCQ treatment also enhanced the antitumor activity of anti-PD-1 antibody in two different low TMB melanoma mouse models [61]. Taken together, these findings suggest that targeting autophagy in TAMs may be a promising approach and may open up the possibility of different drug delivery modalities such as lipid nanoparticles that hone to macrophages, for autophagy drugs.
Autophagy in natural killer (NK) cells, dendritic cells (DCs) and cancer-associated fibroblasts (CAFs)
In addition to T cells and macrophages, there is evidence that inhibiting autophagy enhances the infiltration and cytotoxicity of NK cells in melanoma tumors [69]. Less is known about autophagy in DCs in a cancer context, but Ppt1 deficient DCs were able to cross-present viral antigens more efficiently to T cells, suggesting autophagy inhibition could enhance DCs function in the TME [70]. CAFs present another important part of TME and produce structural components of extracellular matrix like collagen, and growth cytokines. Recent studies indicate that autophagy in CAFs promotes the secretion of interleukin-6 to support tumor growth [19]. In lung cancer, autophagy in CAFs were critical for enabling metastasis [22].
Autophagy modulators for preclinical and clinical trials
Given the evidence that autophagy supports tumor growth and promotes resistance to multiple types of cancer therapy [50-52,71,72], the identification of novel chemical inhibitors or inducers of autophagy is a major unmet need. Many of the proteins involved in autophagy have proven difficult to drug. In addition, despite the evidence provided above, the concern that an autophagy inhibitor will induce tumorigenesis weighs heavily on the field.
Since novel autophagy inhibitors or inducers could produce unknown side effects, they can only be ethically tested in patients with advanced cancer refractory to standard-of-care therapy. These patients are typically not as concerned about the development of polyps as they are about the known cancer that needs to be treated. If novel drugs are not safe or do not produce benefit in the advanced cancer setting, they are unlikely to be developed for other cancer contexts, such as a preventative medication in premalignant disease. The following section will detail efforts to develop novel autophagy inhibitors and their effects on models of advanced cancers.
ULK1/2 inhibitors
An ULK1/2 inhibitor (ULKi) blocks early autophagy and can be a promising therapeutic strategy for cancer. SBI-0206965 has been identified as a potential ULKi that arrests tumor growth in multiple cancer types including non-small cell lung cancer (NSCLC), neuroblastoma, and renal carcinoma [73,74]. SBI-0206965 elevated the sensitivity of daunorubicin in acute myeloid lymphoma [75]. The specificity of SBI-0206965 to ULK1 is questionable, as it has also been found to inhibit focal adhesion kinase (FAK) and AMPK [73]. ULK-100 and ULK-101 are more potent and selective ULKi [76]. ULK-101 suppresses early autophagy induction and induced NSCLC cell death [76]. MRT67307 and MRT68921 are also novel ULKi. They showed in vitro performance and established tumor suppressive activity [61]. DCC-3116, another potent and selective ULKi showed preclinical antitumor activity in combination with MAPKi trametinib. A first-in-human phase 1 study of DCC-3116 monotherapy and with trametinib evaluating the safety, tolerability, clinical activity, pharmacokinetics, and pharmacodynamics in RAS or RAF mutant advanced or metastatic solid tumors patients is ongoing (NCT04892017). This clinical development positions ULK1 as the first autophagy-specific target that has a novel inhibitor in clinical trials.
VPS34 inhibitors
VPS34 is the major lipid kinase that prepares autophagic membranes for the conjugation machinery to be assembled. Targeting VPS34 kinase activity genetically or pharmacologically has a broad impact on the immune landscape of melanoma and colorectal cancer (CRC), by inducing the infiltration of NK cells, CD8+, and CD4+ T effector cells into the TME and turning an immune-cold to an immune-hot TME [60]. In addition, VPS34i (SAR405 or SB02024) treatment in animal models of melanoma or CRC improved the therapeutic efficacy of PD-1 and PD-L1 targeting antibodies [77]. Furthermore, the combination of anti-PD-1/PD-L1 antibodies and VPS34i augments the production of pro-inflammatory cytokines and chemokines such as CCL5, CXCL10, and IFN-γ, as well as the accumulation of CD4+, CD8+ T cells, and NK cells, DCs, and M1 macrophages into the tumor bed enhancing the efficacy of immunotherapy [72]. The major concern with VPS34 inhibitors is that VPS34 can play a role in non-autophagic endocytic trafficking [78], which could lead to off-target toxicity. So far, there is no VPS34 inhibitor that has entered in clinical trials.
V-ATPase inhibitors
V-ATPase is a multi-subunit transmembrane complex that acidifies the lysosomes. While not directly an autophagy target, targeting V-ATPase blocks autophagic flux [79]. 249C is a newly discovered class of dihydro-pyrazole-5-carboxamide compounds that targets the V-ATPase subunit ATP6V1H, inhibiting biochemical activity, lysosomal acidification, and micropinocytosis [80]. The excellent pharmacological properties and tolerability of 249C distinguishes it from other V-ATPase inhibitors such as bafilomycin, which can only be used as a tool compound. Cancer cells expressing KRASG13D and KRASG12V mutations are dependent on V-ATPase and are vulnerable to 249C treatment [80]. If a 249C derivative were to be developed for clinical trials, it would provide an alternative approach to targeting the lysosome in addition to PPT1 inhibitors.
PPT1 inhibitors
PPT1 is a lysosomal hydrolase that cleaves thioester linkages in palmitoylated proteins and facilitates their degradation. PPT1 regulates V-ATPase subcellular localization thereby regulating lysosomal acidification and autophagy [40,81]. Palmitoylation is an important post-transcriptional protein modification involved in protein-protein interactions and subcellular membrane localization of the substrates [82]. Advances in our understanding of CQ and HCQ action led to the identification of PPT1 as an autophagy target. It was previously shown that CQ and HCQ at high concentrations effectively block autophagy in cancer cells yet elicit little cytotoxicity as single agents [83]. However, the molecular target of these drugs was unknown limiting further development. CQ and its derivative HCQ accumulate preferentially in the lysosomes [84]. It was shown that Lys01, a dimeric form of CQ produced a 10-fold more potent autophagy blockade than CQ or HCQ [83]. Recently, leveraging functional knowledge gained by testing Lys01 in preclinical models, dimeric quinacrines [40] and long-linkered dimeric chloroquines [81,85] were generated that were 100-1000-fold more potent than CQ [40,81,85]. These molecules, along with Lys01 and CQ were used to identify PPT1 as their molecular target [40,81]]. With this advance in the understanding of the molecular target of CQ, other novel inhibitors are now in development. For example, PPT1 inhibitor GNS561 has already entered clinical trials [86]. The completed GNS561 phase I trial established the safety of the molecule and a phase 2 combination drug trial of trametinib and GNS561 in KRAS mutant cholangiocarcinoma (NCT03316222) has recently been launched [86,87]. While excitement is growing about novel PPT1 inhibitors, more work needs to be done to identify which patients are most likely to benefit from PPT1 inhibition.
Autophagy inducers
While much effort has been placed on developing autophagy inhibitors, there are proponents of the concept of autophagy induction as a novel approach to cancer therapy [88]. ABTL0812 targets the peroxisome proliferator-activated receptor alpha and gamma (PPARα and PPARγ), Akt/mTOR axis and induces autophagic cell death in tumors [89,90]. ABTL0812 has completed its first-in-human clinical trial with a high safety and efficacy profile (NCT02201823). The recommended dose for the Phase II clinical trial has been determined. In this protocol, ABTL0812 has been administered as first-line therapy in combination with paclitaxel and carboplatin for the treatment of advanced NSCLC patients (NCT03366480)[91]. Although the development of ABTL0812 as an autophagy inducer is intriguing it is likely that the cell death induced by this drug is due to an autophagy-independent mechanism. Given the findings that host cell autophagy can enable tumor growth, there may be an added concern that in some cases autophagy induction could lead to more accelerated growth in advanced cancer.
Resistance mechanisms to autophagy inhibition in cancer
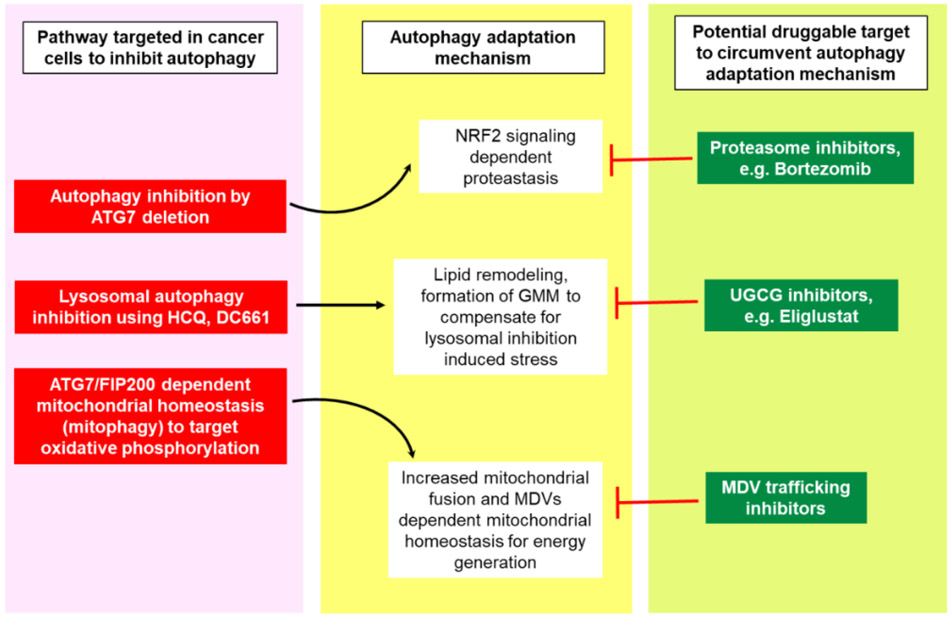
Multiple autophagy inhibitors have been tested in pre-clinical and clinical studies. Except for the pre-clinical data of potent PPT1 inhibitors such as GNS561 and DC661 [81,86,87], there is little evidence to suggest that autophagy inhibitors show single-agent anti-cancer activity. Genetic and pharmacological studies in animal models of cancer suggest that cancer cells can induce complementary pathways to mitigate the effects of autophagy inhibition [92-94] (Figure 3). Targeting such adaptive pathways along with autophagy presents a different approach that can be tested in the clinic. In one study, knock out of some of the twelve essential autophagy genes in multiple autophagy-dependent cancer lines induced cell death in the majority of cells; however, a few clones survived and showed an adaptation to this loss of autophagy [93]. One common mechanism that enabled the survival of these clones was the upregulation of a nuclear factor erythroid 2-related factor 2 (NRF2)-dependent antioxidant response and protein homeostasis [93]. This NRF2 dependency was also seen when autophagosome formation was completely lost during autophagy, by knocking out either upstream or downstream regulator genes of autophagosome formation, RB1CC1/FIP200 and ATG7, respectively [95]. In another study, cancer cell clones which survived the selective pressure of autophagy inhibition were found to be defective in mitophagy [92]. These mitophagy defective clones increased their dependency on mitochondrial fusion, and an autophagy-independent mitochondrial-derived vesicles (MDVs) formation to degrade damaged mitochondria to maintain mitochondrial homeostasis [92]. This report suggested that co-inhibition of autophagy and MDVs trafficking could produce better outcome in cancer than targeting autophagy alone [92].
Figure 3. Autophagy adaptation mechanisms as novel potential targets to augment autophagy inhibitory effects on cancer.
Autophagy inhibition by different strategies can induce compensatory pathways which can mitigate the anti-cancer effects of targeting autophagy in cancer. Combining the inhibitors of autophagy resistance pathways with autophagy inhibiting agents in cancer is a new approach which could be used to augment anticancer effects of autophagy inhibitors as a single agent or in combination with standard of care drugs that induce cytoprotective autophagy. Mitochondrial-derived vesicles (MDVs); GM1 + membrane microdomains (GMM); NRF2, nuclear factor erythroid 2-related factor 2; UGCG, UDP-Glucose Ceramide Glucosyltransferase.
We have reported recently that following treatment with lysosomal autophagy inhibitors (LAI), HCQ or DC661, melanoma cells increased sphingolipid and cholesterol levels that were associated with increased GM1+ membrane microdomains (GMM) in plasma membranes and lysosomes. Targeting UDP-glucose ceramide glucosyltransferase (UGCG), a rate-limiting enzyme for glycosphingolipids synthesis, synergistically augmented LAI induced cytotoxicity in-vitro. Combining the FDA approved UGCG inhibitor eliglustat with LAI produced excellent anti-tumor activity in vivo [94].
Concluding remarks and future perspectives
The tumor cell enabling role of autophagy in cancer suggests that targeting autophagy in cancer is a reasonable approach to boost the efficacy of anti-cancer therapies in advanced cancer. The study of autophagy in cancer has expanded dramatically, but there remains an unmet need to clinically translate these findings. Even though there are a number of HCQ trials and a few novel autophagy inhibitors entering clinical trials, targeting autophagy in patients is still in its infancy as a field. There are multiple challenges and unanswered questions that have so far limited the development of novel autophagy inhibitors and inducers for cancer and other diseases (see outstanding questions).
Outstanding Questions Box.
Do macroautophagy levels vary in different cancer types and thus require different dosage of autophagy inhibitors?
What is the best approach for autophagy modulation at different stages of cancer, either as a single agent or as a combined therapy?
How are different types of selective autophagy programs regulated in specific cancers?
Would it be more effective to target selective autophagy pathways like ferritinophagy compared to broadly targeting autophagy?
Does autophagy inhibition enhance the efficacy of standard of care cancer therapies in the clinic?
Is there a clinical context where an autophagy inhibitor can produce responses as a single agent?
To answer these questions there is a need to develop more potent and specific inhibitors of the pathway. The development of novel tool compounds to study autophagy could help us hone in on the best targets for drug development. Finally, identifying predictive and pharmacodynamic biomarkers that can be translated from the lab into the clinic would speed up clinical development of promising novel autophagy inhibitors for cancer.
Highlights:
Autophagy is a degradative and recycling process that is upregulated in cancer cells.
New advances in understanding the mechanism of autophagy have uncovered novel potential targets for drug development.
Clinical trials involving hydroxychloroquine have demonstrated the safety of targeting autophagy, but efficacy could be improved.
Autophagy regulates tumor immunity.
Novel autophagy inhibitors are entering clinical trials.
Glossary
- AMP-activated protein kinase (AMPK)
AMPK is a heterotrimeric protein complex which can sense intracellular ATP, ADP, and AMP concentrations to maintain cellular energy homeostasis.
- Autophagy-related (ATG) genes/proteins
There are 40 ATG proteins that have been identified in total, of which at least 19 are directly involved in the biogenesis of the autophagosome, including nucleation, elongation, and closure of the phagophore membrane. ATG proteins also have autophagy-independent functions.
- Cargo receptors
Autophagy cargos like organelles and proteins are marked by ubiquitination for their degradation. Ubiquitinated cargos are recognized by their specific cargo receptor proteins (selective autophagy), e.g., sequestosome-1 (SQSTM1/p62), nuclear receptor coactivator 4 (NCOA4), and the neighbor of BRCA1 (NBR1) gene (NBR1), and then recruited to the LC3-II in the autophagosome.
- Major histocompatibility complex (MHC)
MHC is a group of genes that code for a variety of cell surface proteins on antigen presenting cells to display antigen peptides to specific T cell receptors. Major types of MHC molecules are MHC class I and class II that are presented on nucleated cells (almost all cells) and antigen presenting cells (e.g., macrophages, dendritic cells etc.), respectively.
- Mammalian target of rapamycin complex 1 (mTORC-1)
mTORC-1 is a nutrient sensor and growth regulator. Under nutrient sufficient conditions activated mTORC-1 inhibits autophagy and promotes cellular growth.
- Microtubule-associated protein (MAP) light chain 3 (LC3)
LC3 is a human homologue of yeast Atg8, is a crucial part of autophagy. The cytosolic form of LC3 is called LC3-I (18 kDa) and the phagophore membrane-bound form upon autophagy induction is LC3-II (16 kDa). An increase in the ratio of LC3-II/LC3-I serves as one of the markers for autophagosomes formation and can be detected using sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
- Tumor microenvironment (TME)
The tumor microenvironment is the surrounding environment of a tumor that includes immune cells, fibroblasts, blood vessels and the extracellular matrix.
- Unc51-like kinase 1 (ULK1) complex
ULK is the first complex of autophagy pathway and consists of ULK1, ATG13, family interacting protein of 200 kDa (FIP200), ATG13 and ATG101. AMPK inactivates mTORC-1 and activates ULK1 to initiate autophagy process.
- Vacuolar protein sorting 34 (VPS34) complex (also known as Class III phosphatidylinositol 3-kinases (PI3KC3)
A family of lipid kinases that add a phosphate group to the 3′ hydroxyl on the inositol ring of phosphoinositides and generates phosphatidylinositol 3-phosphate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: RKA is inventor on patents related to dimeric chloroquines and quinacrines and founder of Pinpoint Therapeutics.
References:
- 1.Aman Y et al. (2021) Autophagy in healthy aging and disease. Nat Aging 1, 634–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qureshi-Baig K et al. (2020) Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy 16, 1436–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang L et al. (2019) Autophagy inhibition potentiates the anti-angiogenic property of multikinase inhibitor anlotinib through JAK2/STAT3/VEGFA signaling in non-small cell lung cancer cells. J Exp Clin Cancer Res 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew R et al. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang XH et al. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 6.Qu X et al. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laddha S. v. et al. (2014) Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res 12, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaravadi RK (2008) Autophagy-induced tumor dormancy in ovarian cancer. J Clin Invest 118, 3837–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JY et al. (2016) Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev 30, 1704–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataura T et al. (2022) Autophagy promotes cell survival by maintaining NAD levels. Dev Cell 57, 2584–2598.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto K et al. (2020) Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strohecker AM and White E (2014) Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy 10, 384–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang A and Kimmelman AC (2014) Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy 10, 1683–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang A et al. (2018) Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov 8, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karsli-Uzunbas G et al. Autophagy Is Required for Glucose Homeostasis and Lung Tumor Maintenance. DOI: 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huo Y et al. (2013) Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov 3, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strohecker AM et al. (2013) Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 3, 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X et al. (2015) Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov 5, 410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudnick JA et al. (2021) Autophagy in stromal fibroblasts promotes tumor desmoplasia and mammary tumorigenesis. Genes Dev 35, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D et al. (2018) Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nature Communications 2018 9:1 9, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei J et al. (2016) Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 17, 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren Y et al. (2021) Autophagic secretion of HMGB1 from cancer-associated fibroblasts promotes metastatic potential of non-small cell lung cancer cells via NFκB signaling. Cell Death Dis 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noman MZ et al. (2018) Targeting autophagy blocks melanoma growth by bringing natural killer cells to the tumor battlefield. Autophagy 14, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangwala R et al. (2014) Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10, 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.AlMasri SS et al. (2021) Encouraging long-term survival following autophagy inhibition using neoadjuvant hydroxychloroquine and gemcitabine for high-risk patients with resectable pancreatic carcinoma. Cancer Med 10, 7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeh HJ et al. (2020) A randomized phase II preoperative study of autophagy inhibition with high-dose hydroxychloroquine and gemcitabine/nab-paclitaxel in pancreatic cancer patients. Clinical Cancer Research 26, 3126–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeh HJ et al. (2020) A Randomized Phase II Preoperative Study of Autophagy Inhibition with High-Dose Hydroxychloroquine and Gemcitabine/Nab-Paclitaxel in Pancreatic Cancer Patients. Clin Cancer Res 26, 3126–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karasic TB et al. (2019) Effect of Gemcitabine and nab-Paclitaxel With or Without Hydroxychloroquine on Patients With Advanced Pancreatic Cancer: A Phase 2 Randomized Clinical Trial. JAMA Oncol 5, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogl DT et al. (2014) Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy 10, 1380–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinsey CG et al. (2019) Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 25, 620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S et al. (2019) Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev Cell 49, 130–144.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moretti F et al. (2018) TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita K et al. (2018) Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol 217, 3817–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vliet AR et al. (2022) ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Mol Cell 82 [DOI] [PubMed] [Google Scholar]
- 35.Melia TJ et al. (2020) Autophagosome biogenesis: From membrane growth to closure. Journal of Cell Biology 219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao YG et al. (2021) Machinery, regulation and pathophysiological implications of autophagosome maturation. Nature Reviews Molecular Cell Biology 2021 22:11 22, 733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X et al. (2021) New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy 17, 2680–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González-Rodríguez P et al. (2022) SETD2 transcriptional control of ATG14L/S isoforms regulates autophagosome-lysosome fusion. Cell Death Dis 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pechincha C et al. (2022) Lysosomal enzyme trafficking factor LYSET enables nutritional usage of extracellular proteins. Science 378 [DOI] [PubMed] [Google Scholar]
- 40.Rebecca VW et al. (2017) A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov 7, 1266–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratto E et al. (2022) Direct control of lysosomal catabolic activity by mTORC1 through regulation of V-ATPase assembly. Nat Commun 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galluzzi L and Green DR (2019) Autophagy-Independent Functions of the Autophagy Machinery. Cell 177, 1682–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunha LD et al. (2018) LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amaravadi RK et al. (2019) Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov 9, 1167–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boone BA et al. (2015) Safety and Biologic Response of Pre-operative Autophagy Inhibition with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol 22, 4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fei N et al. (2021) SMAD4 loss is associated with response to neoadjuvant chemotherapy plus hydroxychloroquine in patients with pancreatic adenocarcinoma. Clin Transl Sci 14, 1822–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JJ et al. (2021) Clinical Translation of Combined MAPK and Autophagy Inhibition in RAS Mutant Cancer. Int J Mol Sci 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foth M and McMahon M (2021) Autophagy Inhibition in BRAF-Driven Cancers. Cancers (Basel) 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caksa S et al. (2022) The future of targeted kinase inhibitors in melanoma. Pharmacol Ther 239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy JMM et al. (2017) Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ojha R et al. (2019) Er translocation of the mapk pathway drives therapy resistance in BRAF-mutant melanoma. Cancer Discov 9, 396–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma XH et al. (2014) Targeting ER stress–induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 124, 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehnert JM et al. (2022) BAMM (BRAF Autophagy and MEK Inhibition in Melanoma): A Phase I/II Trial of Dabrafenib, Trametinib, and Hydroxychloroquine in Advanced BRAFV600-mutant Melanoma. Clinical Cancer Research 28, 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee CS et al. (2019) MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc Natl Acad Sci U S A 116, 4508–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinsey CG et al. (2019) Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nature Medicine 2019 25:4 25, 620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryant KL et al. (2019) Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 25, 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravichandran M et al. (2022) Coordinated Transcriptional and Catabolic Programs Support Iron-Dependent Adaptation to RAS–MAPK Pathway Inhibition in Pancreatic Cancer. Cancer Discov 12, 2198–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santana-Codina N et al. (2022) NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron–Sulfur Cluster Proteins. Cancer Discov 12, 2180–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain V and Amaravadi RK (2022) Pumping Iron: Ferritinophagy Promotes Survival and Therapy Resistance in Pancreatic Cancer. Cancer Discov 12, 2023–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noman MZ et al. (2020) Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci Adv 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma G et al. (2020) PPT1 inhibition enhances the antitumor activity of anti-PD-1 antibody in melanoma. JCI Insight 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poillet-Perez L et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat Cancer DOI: 10.1038/s43018-020-00110-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poillet-Perez L et al. (2018) Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeVorkin L et al. (2019) Autophagy Regulation of Metabolism Is Required for CD8+ T Cell Anti-tumor Immunity. Cell Rep 27, 502–513.e5 [DOI] [PubMed] [Google Scholar]
- 65.Lawson KA et al. (2020) Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586, 120–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng J et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1-mutant lung cancer. Nat Cancer DOI: 10.1038/s43018-021-00208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto K et al. (2020) Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy 16, 1524–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vitale I et al. (2019) Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab 30, 36–50 [DOI] [PubMed] [Google Scholar]
- 69.Mgrditchian T et al. (2017) Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A 114, E9271–E9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ou P et al. (2019) Thioesterase PPT1 balances viral resistance and efficient T cell crosspriming in dendritic cells. J Exp Med 216, 2091–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mele L et al. (2020) The role of autophagy in resistance to targeted therapies. Cancer Treat Rev 88, 102043. [DOI] [PubMed] [Google Scholar]
- 72.Noman MZ et al. (2020) Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci Adv 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egan DF et al. (2015) Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell 59, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dower CM et al. Targeted Inhibition of ULK1 Promotes Apoptosis and Suppresses Tumor Growth and Metastasis in Neuroblastoma. DOI: 10.1158/1535-7163.MCT-18-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiu L et al. (2020) Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci 243 [DOI] [PubMed] [Google Scholar]
- 76.Petherick KJ et al. (2015) Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem 290, 11376–11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janji B et al. (2020) Firing up the cold tumors by targeting Vps34. Oncoimmunology 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jaber N et al. (2016) Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J Cell Sci 129, 4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mauvezin C and Neufeld TP (2015) Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11, 1437–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tolani B et al. (2022) Ras-mutant cancers are sensitive to small molecule inhibition of V-type ATPases in mice. Nature Biotechnology 2022 10, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rebecca VW et al. (2019) PPT1 Promotes Tumor Growth and Is the Molecular Target of Chloroquine Derivatives in Cancer. Cancer Discov 9, 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang MM and Hang HC (2017) Protein S-palmitoylation in cellular differentiation. Biochem Soc Trans 45, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McAfee Q et al. (2012) Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A 109, 8253–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Homewood CA et al. (1972) Lysosomes, pH and the anti-malarial action of chloroquine. Nature 235, 50–52 [DOI] [PubMed] [Google Scholar]
- 85.Wang Y et al. (2021) Anticancer properties of bisaminoquinolines with modified linkers. Bioorg Med Chem Lett 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brun S et al. (2021) GNS561, a New Autophagy Inhibitor Active against Cancer Stem Cells in Hepatocellular Carcinoma and Hepatic Metastasis from Colorectal Cancer. J Cancer 12, 5432–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brun S et al. (2021) GNS561, a clinical-stage PPT1 inhibitor, is efficient against hepatocellular carcinoma via modulation of lysosomal functions. 10.1080/15548627.2021.1988357 18, 678–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mariño G et al. (2014) Two distinct self-destructive processes, autophagy ('self-eating') (BOX 1) and apoptosis ('self-killing'). DOI: 10.1038/nrm3735 [DOI] [Google Scholar]
- 89.Felip I et al. (2019) Therapeutic potential of the new TRIB3-mediated cell autophagy anticancer drug ABTL0812 in endometrial cancer. Gynecol Oncol 153, 425–435 [DOI] [PubMed] [Google Scholar]
- 90.Erazo T et al. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. Clin Cancer Res 22 [DOI] [PubMed] [Google Scholar]
- 91.López-Plana A et al. (2020) The novel proautophagy anticancer drug ABTL0812 potentiates chemotherapy in adenocarcinoma and squamous nonsmall cell lung cancer. Int J Cancer 147, 1163–1179 [DOI] [PubMed] [Google Scholar]
- 92.Towers CG et al. (2021) Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy. Dev Cell 56, 2029–2042.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Towers CG et al. (2019) Cancer Cells Upregulate NRF2 Signaling to Adapt to Autophagy Inhibition. Dev Cell 50, 690–703.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jain V et al. (2022) Targeting UGCG overcomes resistance to lysosomal autophagy inhibition. Cancer Discov DOI: 10.1158/2159-8290.CD-22-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Towers CG et al. (2020) Autophagy-dependent cancer cells circumvent loss of the upstream regulator RB1CC1/FIP200 and loss of LC3 conjugation by similar mechanisms. Autophagy DOI: 10.1080/15548627.2020.1741204/SUPPL_FILE/KAUP_A_1741204_SM2573.ZIP [DOI] [PMC free article] [PubMed] [Google Scholar]