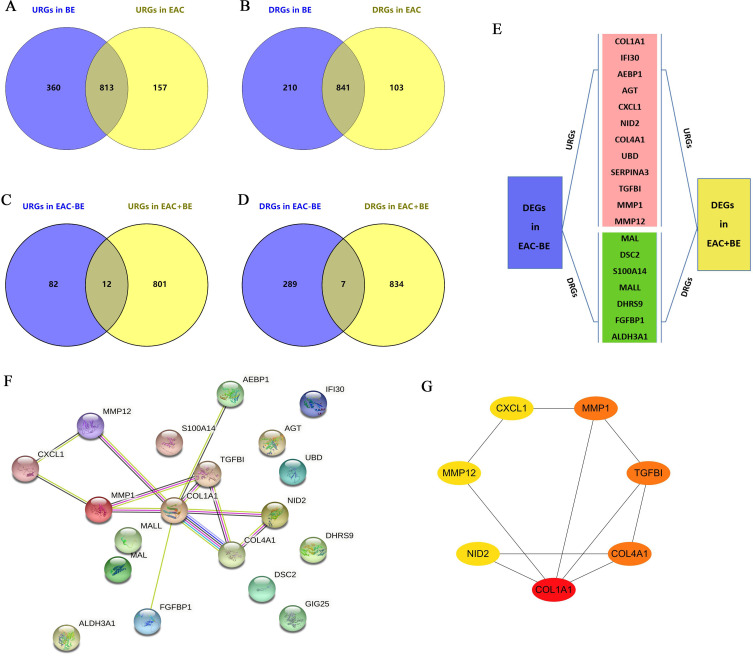
Figure 2.
Exploration of hub genes during the development of NE-BE-EAC. (A and B) Relative to NE, up- and down-regulated DEGs overlapping in BE and EAC were identified by Wayne analysis. (C and D) Compared to BE, the upregulated and downregulated genes in EAC are labeled as “URGs in EAC-BE” and “DRGs in EAC-BE”, respectively. Relative to NE, overlapping up- and down-regulated genes in BE and EAC were labeled as “URGs in EAC+BE” and “DRGs in EAC+BE”, respectively. Overlapping genes between “URGs in EAC-BE” and “URGs in EAC+BE”, “DRGs in EAC-BE” and “DRGs in EAC+BE” were identified by Wayne analysis. (E) These overlapping DEGs include 12 upregulated genes and 7 downregulated genes. (F) Analysis of protein interactions between these 19 DEGs using the STRING database. (G) Finally, 7 hub genes were screened based on protein interactions using the cytohubba plugin in Cytoscape software. The linkages between the proteins indicate the predicted interactions.