Figure 6.
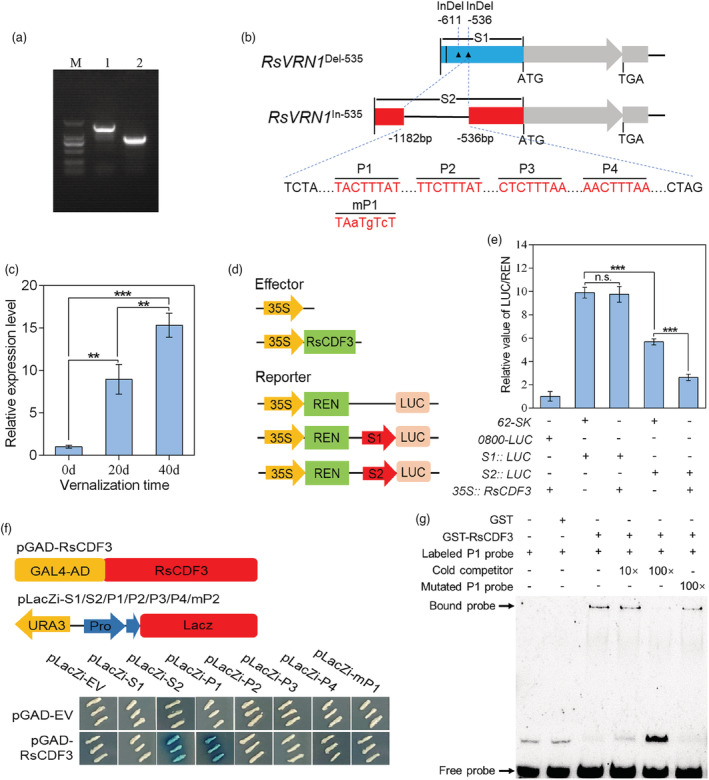
RsCDF3 can directly binds to the promoter of RsVRN1 In‐536 allele. (a) PCR‐based cloning of the RsVRN1 promoter from ‘NAU‐LB’ and ‘Xin‐li‐mei’ genotype, respectively. M: DL2000 marker. 1: NAU‐LB; 2: Xin‐li‐mei. (b) Schematic diagram of the RsVRN1 Del‐536 and RsVRN1 In‐536 promoter fragments used for construction of the transient expression vector. The vertical line and triangle represent one SNP and two InDels in the RsVRN1 promoter of Xin‐li‐mei compared with NAU‐LB, respectively. Four potential DOF binding elements (P1, P2, P3 and P4) within the 647‐bp insertion are indicated in red. The mutated nucleotides are presented in lowercase in mP1 fragment sequences. (c) The expression profile of the RsVRN1 gene during prolonged vernalization period in ‘NAU‐LB’ genotype. (d) Schematic diagrams of the LUC, S1 and S2 reporter constructs used for transient expression assay. (e) Transient expression assays of different promoter fragments from two RsVRN1 genotypes. (f) Yeast one‐hybrid assays showing that RsCDF3 binds to P1 fragment within the 647‐bp insertion. The prey and bait vectors used for the assays are indicated at the top. (g) Analysis of RsCDF3 binding to the P1 fragment of the 647‐bp insertion in an EMSA system. Biotin‐labelled probes were incubated with GST or GST‐tagged RsVRN1. 10× and 100× unlabelled competitor fragments were added to evaluate binding specificity. Values are mean ± SD from three independent biological replicates. Asterisks indicate statistically significant differences using two‐sided Student's t test (**P < 0.01; ***P < 0.001).
