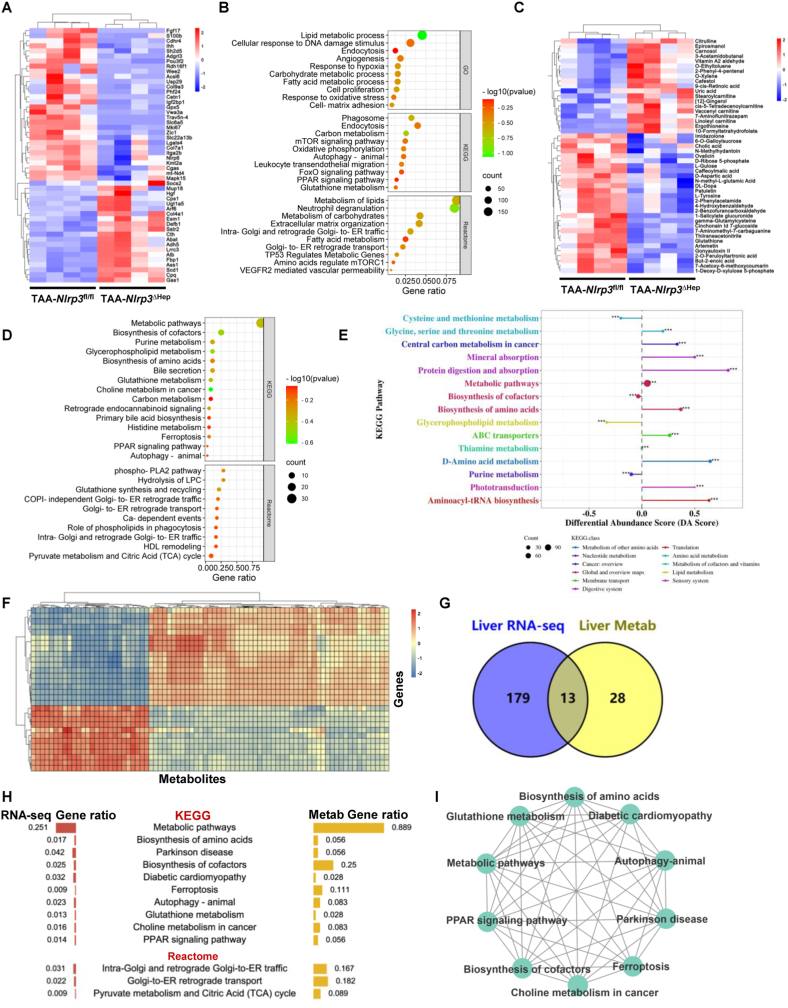
Fig. 6.
Hepatocyte-specific Nlrp3 deletion attenuates liver fibrosis via metabolic reprogramming
(A-E) RNA-sequencing and metabolomics of livers from Nlrp3fl/fl and Nlrp3ΔHep mice injected with TAA for 8 weeks (n = 4/group). RNA-sequencing was performed to show the heatmap (A) and the enriched signaling pathways (B) of DEGs. Ultra HPLC-based metabolomics of livers was applied to reveal the heatmap (C), the enriched signaling pathway (D), and the KEGG metabolic pathway-based differential abundance analysis (E) of differential metabolites. (F–I) Integrative multiomics analysis of the transcriptome and metabolomics. Hierarchical clustering for canonical correlation analysis of the mRNAs and metabolomics (F). Venn diagram (G) and ontology analysis (H) displaying the shared signaling pathways between the transcriptome and metabolomics. Functional enrichment analysis was then performed to gain further insights into the KEGG of DEGs and metabolites (I).