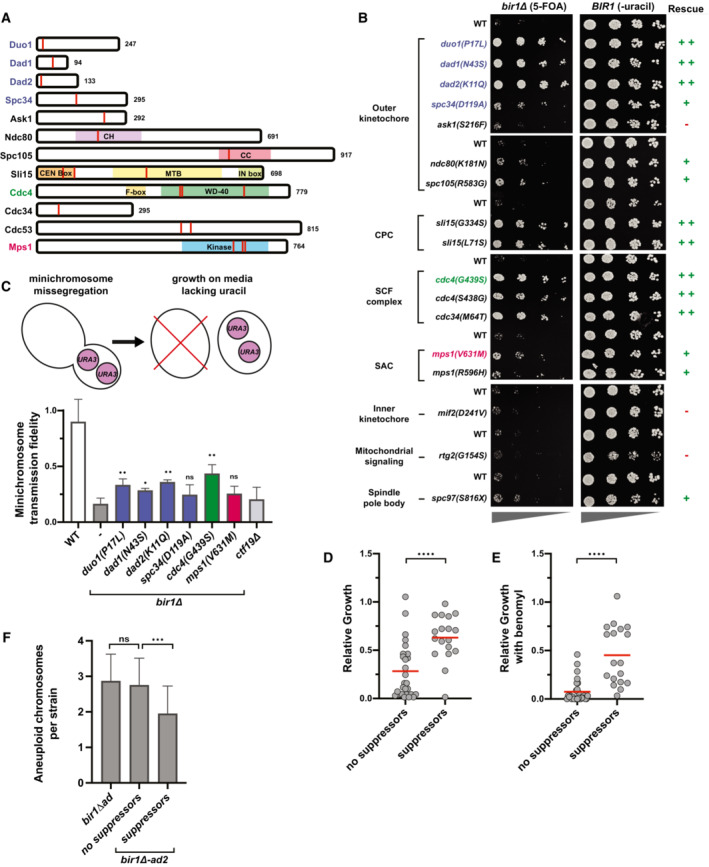
Figure 2. Four main categories of mutations rescue BIR1 deletion.
-
ASchematics showing the location of mutations from Table 1 (red lines). Domains of interest were identified using the Saccharomyces Genome Database.
-
BSerial dilutions of strains engineered with the indicated mutations identified in bir1∆‐ad2 strains were tested for rescue of BIR1 deletion. Ten‐fold serial dilutions on the indicated media are shown. The mutations colored in blue, green, or red were selected for additional characterization.
-
CTop: schematic summarizing how loss of the URA3‐containing plasmid is used to measure relative missegregation rates. Bottom: Proportion of colonies that grow on plates with restrictive (lacking uracil) versus permissive (YPAD) media. Many of the suppressor mutants significantly increase the segregation fidelity of the minichromosome in a bir1∆ background. ctf19∆ serves as a positive‐control for decreased minichromosome transmission fidelity. Statistical significance is relative to bir1∆ alone for three independent experiments. Means and standard deviations are shown.
-
D, EGrowth comparisons of bir1∆‐ad2 strains that contain (18 strains) or do not contain (28 strains) confirmed suppressor mutations as measured by area of colony growth after serial dilution. The mean (red line) is shown. 10‐fold serial dilutions were made on YPAD plates containing either 0.1% DMSO (D) or 10 μg/ml of benomyl (E).
-
FComparison of the mean number of aneuploid chromosomes per strain for bir1∆‐ad2 with and without identified suppressor mutations.
Data information: (ns) not significant; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001; unpaired t‐test.
