Figure 3. Suppressor mutations in the Dam1 complex result in elevated spindle assembly checkpoint activity and minichromosome missegregation.
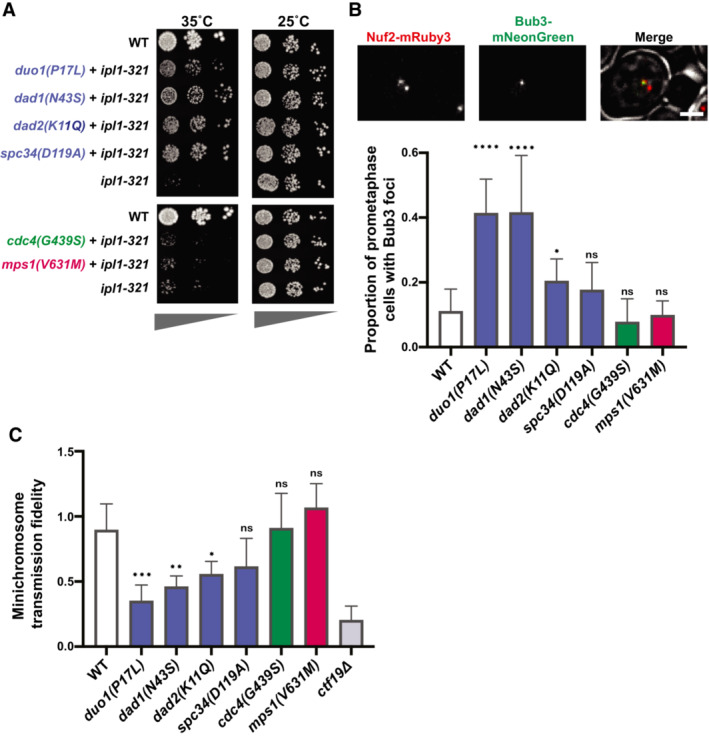
- Serial dilutions on YPAD media at the permissive (25°C) or restrictive (35°C) temperatures for the ipl1‐321 mutation. The suppressor mutations from the Dam1c (blue) were the only mutants that rescued viability at the restrictive temperature.
- Top: representative images showing Bub3‐mNeonGreen localization to kinetochores (Nuf2‐mRuby3), indicating spindle assembly checkpoint activation in wild‐type yeast. Scale bar is 2 μm. Bottom: quantification of the proportion of prometaphase cells with Bub3‐mNeonGreen foci. Only the mutations in Dam1 complex (blue) show a significant increase in SAC activity. Data are from at least five independent experiments. Means and standard deviations are shown.
- Proportion of colonies that grow on plates with restrictive (lacking uracil) versus permissive (YPAD) media after 24 h growth under permissive conditions with a URA3‐containing plasmid. The suppressor mutations in the Dam1c (blue) show a significant reduction of minichromosome segregation fidelity. Data are from three independent experiments. Means and standard deviations are shown.
Data information: (ns) nonsignifcant; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001; unpaired t‐test.
