Figure 4. Suppressor mutations in the Dam1 complex have phenotypes similar to the dam1(S20D) phosphomimic mutation.
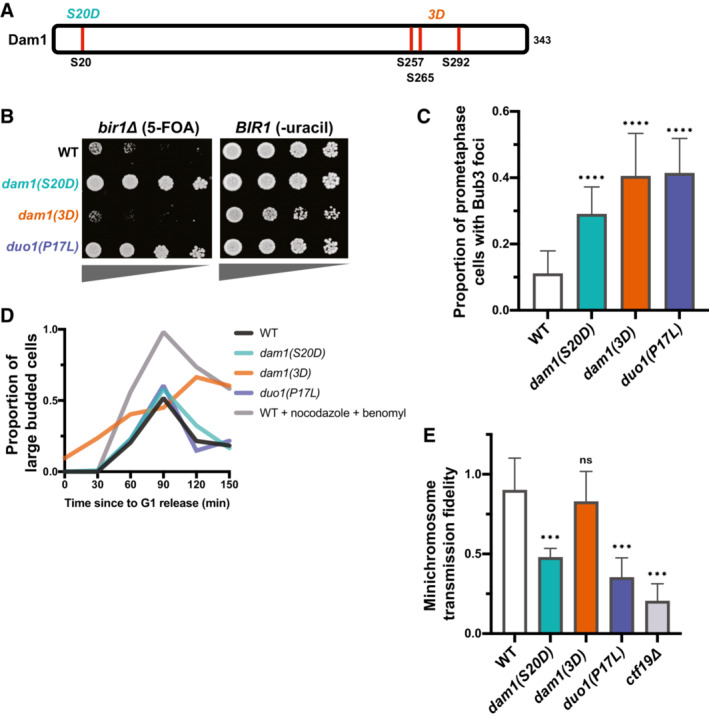
- A schematic showing the relative locations of the Ipl1 phosphosites in Dam1 that are mutated to aspartic acid in the phosphomimics.
- Serial dilutions of strains engineered with the indicated mutations were tested for rescue of BIR1 deletion. Ten‐fold serial dilutions on the indicated media are shown. dam1(S20D) rescues loss of CPC activity to a similar extent as a suppressor mutant in Duo1.
- Quantification of the proportion of prometaphase cells with Bub3‐mNeonGreen foci localized to kinetochores (Nuf2‐mRuby3), indicating SAC activation in all three mutant strains. Data are from at least five independent experiments. Means and standard deviations are shown.
- Degree of cell cycle delay as measured by percentage of large budded cells over time after release from G1 arrest. Wild‐type (WT) cells treated with nocodazole and benomyl were used as a positive control for mitotic arrest.
- Proportion of colonies that grow on plates with restrictive (lacking uracil) versus permissive (YPAD) media after 24 h growth under permissive conditions with a URA3‐containing plasmid. Minichromosome segregation fidelity is reduced in the dam1(S20D) strain, similar to the duo1(P17L) strain. Data are from three independent experiments. Means and standard deviations are shown.
Data information: (ns) nonsignifcant; (***) P < 0.001; (****) P < 0.0001; unpaired t‐test.
