Figure EV2. CRISPR‐Cas9 mediated mutation of outer kinetochore genes and suppression validation.
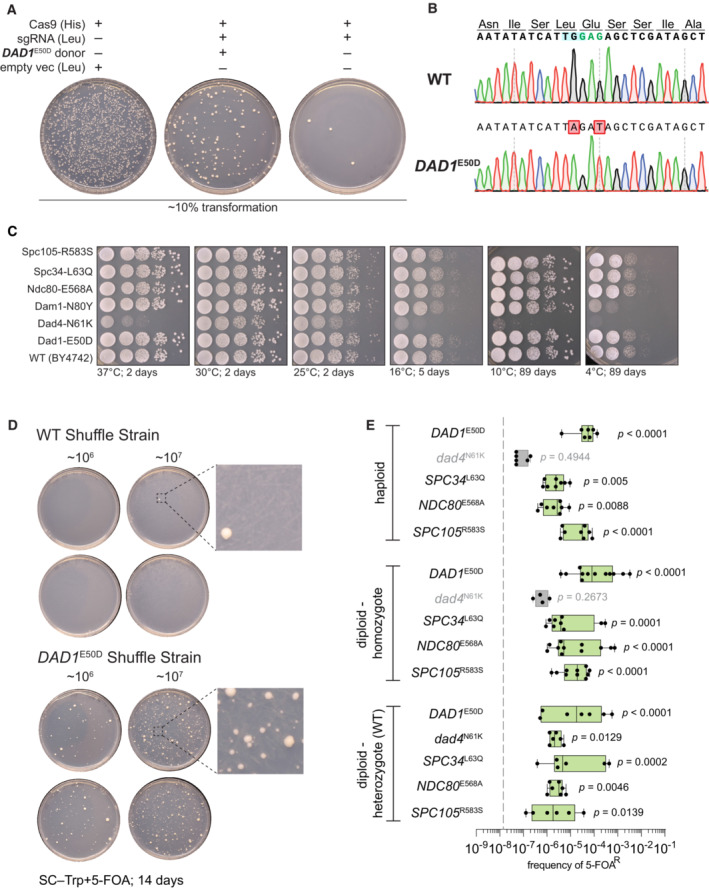
- Example CRISPR/Cas9 editing transformation to scarlessly introduce each point missense mutation to an isogenic histone shuffle strain. Note in the absence of dsDNA donor the sgRNA targeting DAD1 results in a severe killing phenotype and upon co‐transformation with a proper dsDNA, the killing phenotype is rescued.
- Example Sanger tracks for the edited DAD1 E50D mutation and wild‐type sequence. The targeting PAM is highlighted in blue, the edited codon in green, and the modified base pairs in red.
- Growth assay on rich medium (YPD) for the indicated histone shuffle strains with yeast histones. Spots are 10‐fold serial dilutions from starting OD600 of 1.0.
- Example suppressor mutation sufficiency histone‐humanization experiment is shown for the DAD1 E50D mutation. Two concentrations of cells were plated (106 and 107 ml−1).
- Histone humanizations for all tested suppressor mutations are shown. The average rates for wild‐type strains are plotted as a gray‐dashed line. Significance was determined with a Kruskal–Wallis test of the mean frequency of 5‐FOAR for each mutant versus the mean frequency of 5‐FOAR of wild type, with multiple‐comparison corrections with the false discovery rate method. Green boxes represent suppressors who significantly increased the rate of humanization above wild‐type level. Each dot represents a biological replicate of the histone humanization assay (n ≥ 4), the central band represents the median, the box extends from the 25th to 75th percentile, and the whiskers represent the minimum and maximum.
