Figure 6. Molecular basis for suppression of histone humanization by DAD1 E50D and DAM1 N80Y .
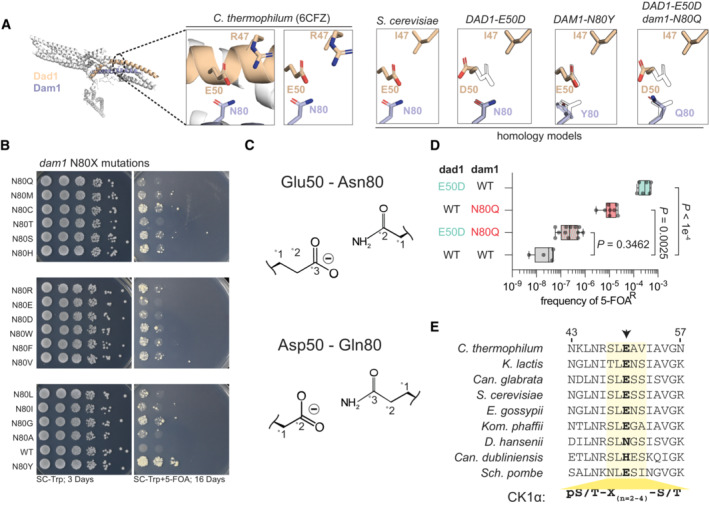
- Homology models of S. cerevisiae DASH/Dam1c complex. The decametric DASH/Dam1c Cyro‐EM structure of C. thermophilum is shown, with Dad1 and Dam1 highlighted. The corresponding homology model is shown to the right, with various mutant models shown (with outlined WT residues for reference). Note the relative positioning of Dad1 residue 50 and Dam1 residue 80.
- Histone‐humanization assay for various dam1 N80X mutants. 5‐FOA is used to counter‐select the yeast histone plasmid, forcing growth with the human histone plasmid. Yeast was serially diluted from a starting culture of 1.0 OD600.
- Cartoon interactions for complementary pairings of Dad1 residue 50 and Dam1 residue 80.
- The dual mutant Dad1‐Asp50—Dam1‐Gln80 DASH/Dam1c fails to humanize, while the single mutants readily humanize. The significance of the mean difference in 5–FOAR frequency was determined with the Mann–Whitney test. Each dot represents a biological replicate (n ≥ 4), the central band represents the median, the box extends from the 25th to 75th percentile, and the whiskers represent the minimum and maximum.
- Protein alignment of Dad1 orthologs, highlighting a conserved casein 1 kinase consensus phosphorylation site (yellow‐shaded region). Arrow indicates the mutant residue 50 of Dad1.
