Figure 1. RNA m6A‐directed formation of heterochromatin in ES cells.
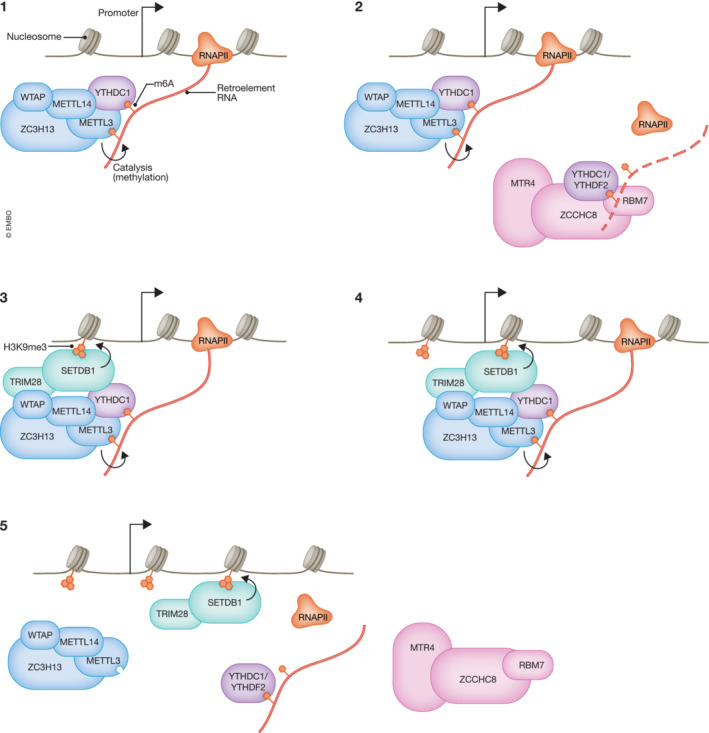
Genomic loci of retrotransposons such as LINE1 and IAP elements are usually decorated by the H3K9me3 and H4K20me3 chromatin marks (Mikkelsen et al, 2007). The former mark is often required for the deposition of the latter, as well as for efficient chromatin silencing (Schotta et al, 2004). N6‐adenine methylation (m6A) on chromatin‐associated retrotransposon RNAs can alter heterochromatic landscape and lead to transcriptional silencing. (1) At first, m6A is deposited on nascent retrotransposon RNAs by the METTL3‐METTL14 complex toward the 5′ end of chromatin‐associated RNAs (Xu et al, 2021). Different m6A readers recognize this modification and mediate various functions, however, the full complexity of their network remains elusive. One such m6A reader YTHDC1, seems to positively regulate METTL3‐METTL14 complex formation. (2) YTHDC1 as well as YTHDF2 are also involved in targeted degradation of the m6A labeled RNAs via the Nuclear Exosome Targeting Complex (NEXT; Liu et al, 2020a; Chelmicki et al, 2021; Chen et al, 2021; Garland et al, 2022). (3) m6A on such RNAs can also lead to METTL3‐dependent recruitment of the histone methyltransferase complex SETDB1‐TRIM28. (4, 5) Subsequent spreading of H3K9me3 mark on proximal nucleosomes results in decreased chromatin accessibility and transcriptional silencing (Liu et al, 2020a; Xu et al, 2021).
