Figure 6. Stat5‐mediated assembly of Ets1/Runx1 enhanceosome.
-
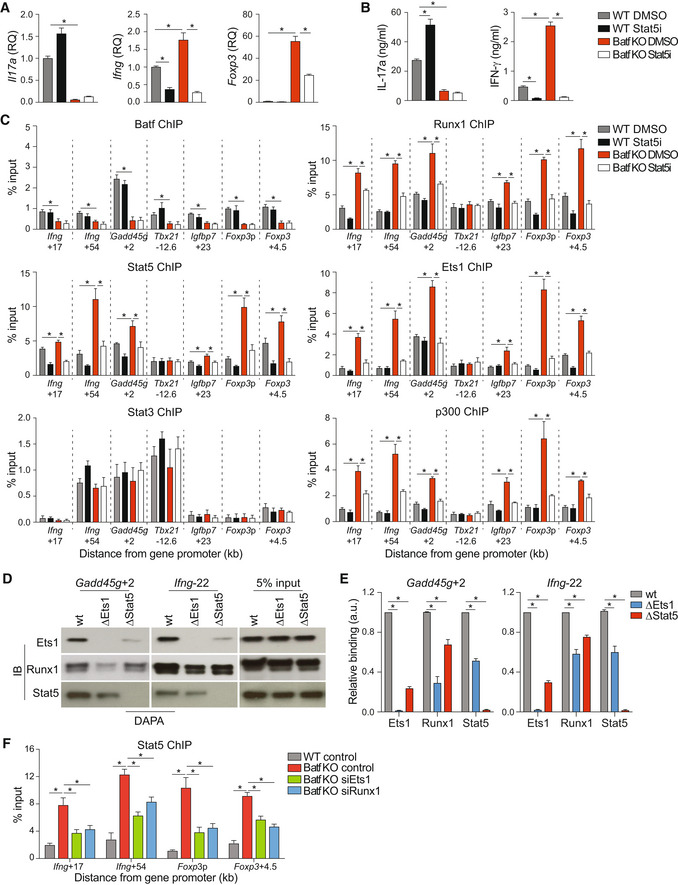
A–CNaïve WT and Batf KO CD4+CD62Lhi T cells cultured under Th17‐polarizing conditions were treated with or without 1 μM Stat5 inhibitor. On day 5, cells were harvested and used for RNA extraction to assess gene expression (A; data were normalized to WT DMSO; Foxp3, unstimulated; Il17a and Ifng, 6 h 2 μg/ml anti‐CD3 restimulation) or restimulated with 2 μg/ml anti‐CD3 for 24 h to assess cytokine production by ELISA (B). WT and Batf KO Th17‐polarized cells were collected on day 5 of culture and used to perform ChIP using antibodies to Stat5, Batf, Ets1, Runx1, Stat3, and p300 (C).
-
D, ENuclear extracts from WT Th17 cells were incubated with biotinylated oligonucleotides containing Ets1‐Runx1‐Stat5‐specific binding sites at the indicated genomic regions (wild‐type or mutant [Δ] as indicated). Immunoblots of precipitated proteins (D), with densitometry measurements (E).
-
FNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 3, cells were transfected with control or siRNA targeting Ets1 or Runx1, cultured for an additional 24 h without restimulation, and used to assess Stat5 binding at the indicated genomic regions by ChIP–qPCR.
Data information: Data are mean ± s.e.m. of four independent experiments. (*P < 0.05; two‐sided t‐test or one‐way ANOVA followed by Tukey's test). a.u., arbitrary unit.
