Figure 2.
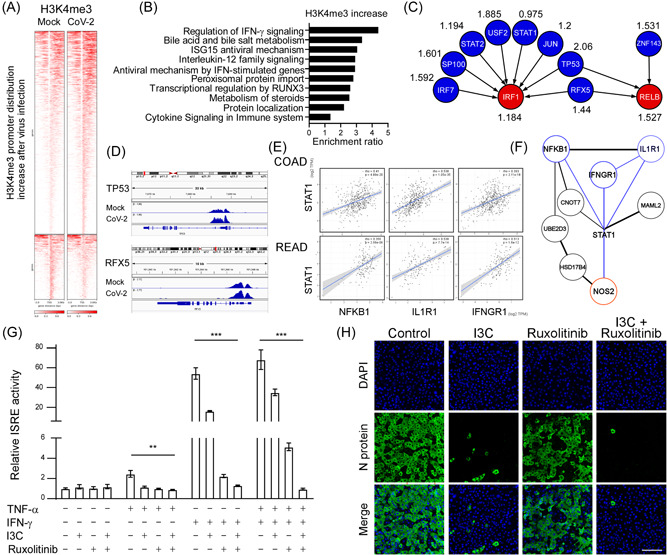
SARS‐CoV‐2 epigenetic activates NF‐κB/IFN‐γ mediated cytokine production and viral infectivity. (A) Heatmap of H3K4me3 ChIP‐sequencing density in a ±3 kb transcription start site region around the summit of each differentially accessible peak in control and virally infected groups in Caco‐2 cell line. (B) Top 10 gene ontology analysis of biological pathways of genes with increased H3K4me3 using WebGestalt (WEB‐based Gene Set Analysis Toolkit) (http://www.webgestalt.org/). (C) The normalized highest H3K4me3 tag intensity in TO‐GCN L1 genes (IRF1 and RELB) and L2 genes (IRF7, SP100, STAT2, USF2, STAT1, JUN, TP53, RFX5, and ZNF143) near promoter regions after being infected with SARS‐CoV‐2 for 48 h compared with mock. The higher normalized value, the more H3K4me3 bind to the promoter region after SARS‐CoV‐2 infection. (D) The Integrative Genomics Viewer browser of H3K4me3 occupancy in TP53, and RFX5 after being infected with SARS‐CoV‐2 for 48 h. (E) Spearman correlation analysis of STAT1 with NFKB1, ILIR1, and IFNGR1 in The Cancer Genome Atlas colon cancer (COAD; n = 460 and rectum cancer data set (READ; n = 171). (F) STRING protein‐protein interaction network showing NFKB1, ILIR1, and IFNGR1 were upstream STAT1/NOS2 axis genes. (G) Luciferase activity fold induction in HEK293T cells transfected with the ISRE reporter plasmid pretreated with 10 μM in I3C, or 4 μM Ruxolitinib for 1 h, then treated with 10 ng/ml human TNF‐α, or 10 ng/ml IFN‐γ for 48 h. Statistical method: two‐way analysis of variance, Tukey post hoc tests, **p < 0.01, ***p < 0.001. (H) Representative immunofluorescence images of the NF‐κB inhibitor (I3C; 10 μM) and the IFN‐γ inhibitor (Ruxolitinib; 4 μM) affect the expression of viral N protein after SARS‐CoV‐2 infection for 48 h in Vero E6 cells. Scale bar = 100 μm. ChIP, chromatin immunoprecipitation; IFN‐γ, interferon‐γ; IRF1, interferon regulatory factor 1; I3C, indole‐3‐carbinol; NF‐κB, nuclear factor‐κB; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; TNF‐α, tumor necrosis factor‐α; TO‐GCN, time‐ordered gene coexpression network.
