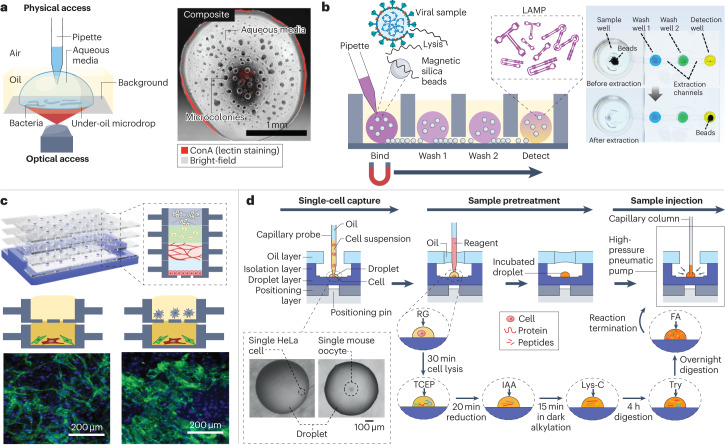
Fig. 7. Cell analysis in open droplet systems.
Open droplet microfluidic platforms enable more flexibility and control in cell culture experiments, and many of the platforms are compatible with conventional in vitro experimental readouts. a, Left: a schematic of an under-oil open microfluidic system used to study the growth dynamics and microcolony motility of wild-type Flavobacterium johnsoniae with physical and optical access. Right: a composite bright-field and fluorescent Concanavalin A (ConA) lectin-stained image of a microdrop obtained with this system 12 h after the culture growth was initiated. b, Left: a schematic of the oil-immersed lossless total analysis system (OIL-TAS) used to perform integrated RNA extraction and detection of SARS-CoV-2. The detection step uses loop-mediated isothermal amplification (LAMP). Right: images of a unit of the OIL-TAS device before and after extraction in which the wells were coloured for visualization. c, Top: a reconfigurable multilayer suspended cell-culture system (‘stacks’) for studying spatiotemporal paracrine signalling across different culture microenvironments. Bottom: immunocytochemistry images show the difference in morphology of the endothelial structures (green) in the endothelial–fibroblast mixture layer when macrophages are absent (left) and present (right) in the top layer. d, Single-cell proteomic analysis using a nanolitre-scale oil–air–droplet chip with a self-aligning monolithic device. The pretreatment of the sample involves the addition of RapiGest (RG), tris(2-carboxyethyl)-phosphine (TCEP), iodoacetamide (IAA), endoproteinase Lys-C, trypsin (Try) and formic acid (FA) to extract proteins from a single cell and digest them into peptides. The sample is then injected into a capillary liquid chromatography column for subsequent liquid chromatography tandem mass spectrometry analysis. The inset shows microscopy images of droplets containing a single cell. Part a is adapted from ref. 31 under a Creative Commons licence CC BY 4.0. Part b is adapted from ref. 33 under a Creative Commons licence CC BY 4.0. Part c is reprinted from ref. 3, Springer Nature Limited. Part d is adapted from ref. 54, American Chemical Society.