Figure 4.
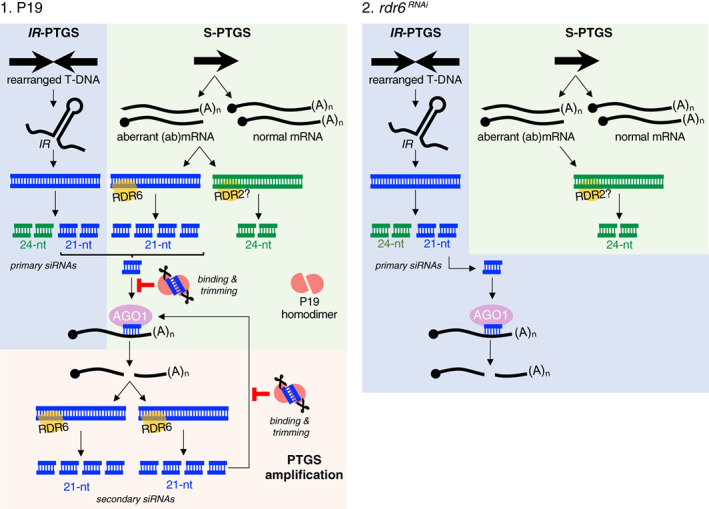
Proposed model for the multilayered action of P19 (scenario 1) during transient gene expression in Nicotiana benthamiana leaves as compared to the effect of RDR6 knockdown (scenario 2) using the rdr6 RNAi background.
In scenario 1, IR‐PTGS involves transgene arrays organized as inverted repeats (IRs). Upon transcription, these form RDR‐independent sources of dsRNA, which is processed into 21‐nt and 24‐nt siRNA species. An additional source of dsRNA is provided through S‐PTGS via RDR6 (leading to 21‐nt siRNA production) and possibly RDR2 (leading to 24‐nt siRNA production), using sense polyA− and uncapped (collectively referred to as to ‘aberrant’) RNAs as templates. 21‐nt siRNAs produced by both IR‐ and S‐PTGS are then loaded into AGO1 to guide endocleavage of complementary sense transgene mRNAs. The ensuing polyA− and uncapped RNA breakdown products being aberrant, they serve, in turn, as novel sources of RDR6‐dependent dsRNA and secondary 21‐nt siRNAs. This RDR6‐amplified phase would contribute the main siRNA bulk in infiltrated tissues, explaining the disproportionally high representation of 21‐nt RNAs. Co‐expressed P19 binds to and causes trimming of both primary and secondary 21‐nt siRNAs. Sequestration of the former, in particular, impedes PTGS amplification by RDR6, resulting in a substantial decrease of the dominant 21 nt siRNA fraction. The 24‐nt siRNA levels remain largely unchanged because they are not bound by P19 and, hence, not trimmed. In scenario 2, reduced RDR6 activity in the rdr6 RNAi background impedes the onset of amplified PTGS, leading to mostly primary (i.e., non‐amplified) 21‐nt siRNA accumulation. The levels of RDR2‐dependent 24‐nt siRNAs presumably accompanying S‐PTGS remain unchanged. While considerably reduced in levels as in scenario 1, the 21‐siRNAs would not undergo trimming.
