Figure 2.
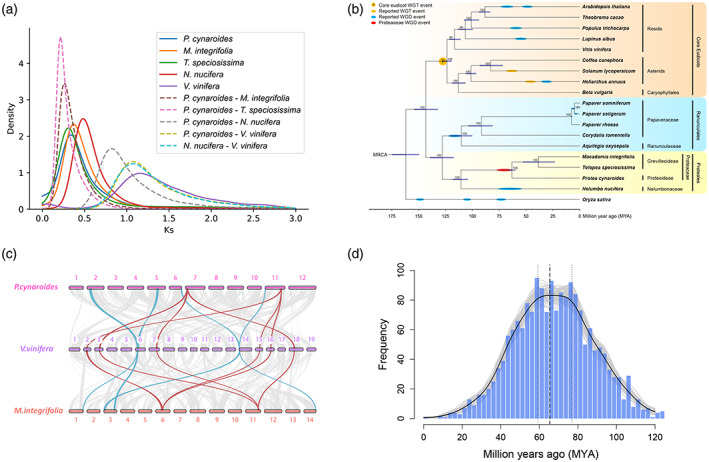
The whole‐genome duplication (WGD) event in Proteaceae.
(a) Ks distributions for anchor pairs (retained paralogs from the WGD event) within Protea cynaroides, Macadamia integrifolia and Telopea speciosissima genome, and for orthologs of P. cynaroides and related species.
(b) Inferred phylogenetic tree with 176 single‐copy genes in 17 species except for Papaver somniferum and Papaver setigerum identified by OrthoFinder. Timing of Proteaceae WGD was estimated in this study, and previously reported whole‐genome triplication (WGT)/WGD events are superimposed on the tree.
(c) Intergenomic co‐linearity among P. cynaroides, Vitis vinifera and M. integrifolia.
(d) Absolute dating of the P. cynaroides WGD event. The age distribution was obtained by phylogenomic dating of P. cynaroides paralogs. The solid black line represents the Kernel Density Estimation (KDE) of the dated paralogs, while the vertical dashed black line represents its peak at 67.9 MYA, which was used as the consensus WGD age estimate. The gray lines represent density estimates from 2500 bootstrap replicates, and the vertical black dotted lines represent the corresponding 90% confidence interval (CI) for the WGD age estimate, 59.267–76.85 MYA (see Experimental Procedures section). The histogram shows the raw distribution of dated paralogs.
