Fig. 6.
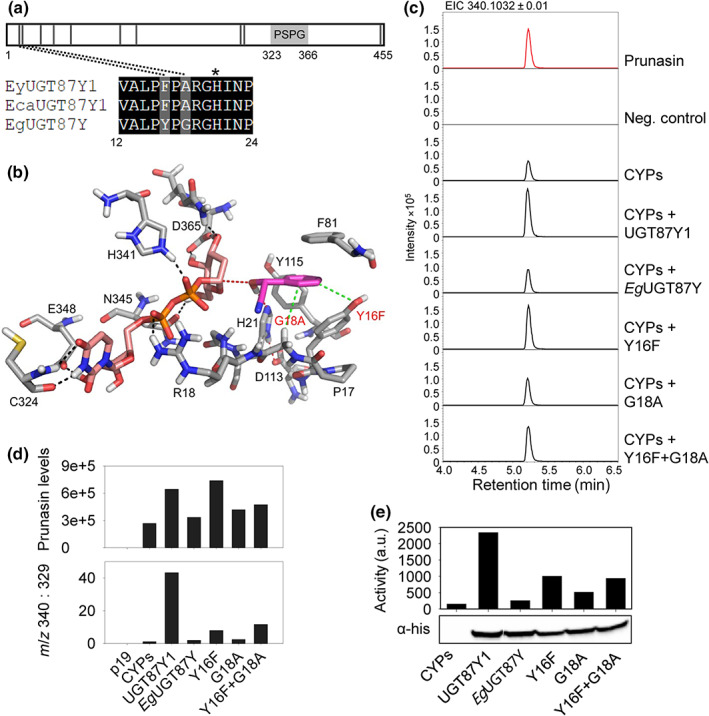
Site‐directed mutagenesis of EgUGT87. (a) Schematic of the UGT87 amino acid sequence indicating the positions of amino acids (black vertical lines) that differ between UGT87Y1 and EgUGT87Y. The PSPG motif is marked with a light gray box. The catalytic His is marked with an asterisk. (b) Active site of EgUGT87 (Eucgr.B03993) (grey carbon atoms) with docked mandelonitrile (magenta carbon atoms) and UDP‐glucose (pink carbon atoms). Nitrogen, oxygen, sulfur and phosphorus atoms are shown in blue, red, yellow and orange, respectively. The benzene moiety of the substrate is mainly recognized by hydrophobic interactions with Y16, F81, and Y115. Mutations of Y16F and G18A (red text) enhance the hydrophobic interactions (green dotted lines) with the benzene ring of mandelonitrile. The catalytic dyad H21 ‐ D113 is shown (red dotted lines) and hydrogen bonds indicated (black dotted lines). (c) Representative extracted ion chromatograms (EIC) corresponding to prunasin (m/z 340.1032 [M + FA]−) of leaf extracts from Nicotiana benthamiana plants transiently expressing Eucalyptus UGTs together with the prunasin CYPs from Eucalyptus yarraensis. (d) Prunasin accumulation in N. benthamiana leaf extracts measured by peak area integration of prunasin (m/z 340; upper panel), and prunasin levels normalized to the pathway derivative putatively annotated as benzoic acid glucoside (m/z 329), shown here as a ratio (m/z 340 : 329; lower panel). (e) In vitro assays using protein extracts prepared from N. benthamiana leaves transiently expressing prunasin CYPs and the UGT variants (top) and Western blot of the His‐tagged UGTs in the protein extract that was used for activity assay (bottom). The UGT activity is plotted as the intensity of the prunasin signal on a TLC plate measured by phospho‐imaging.
