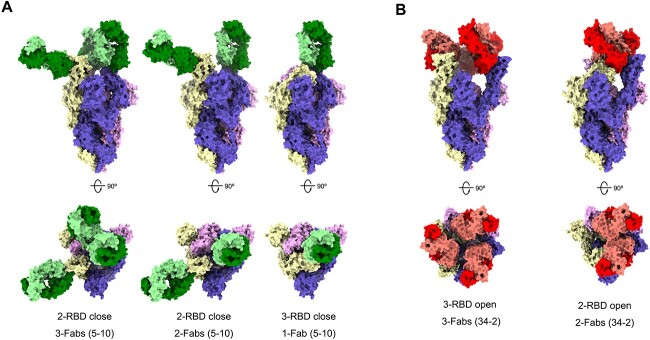
Figure 4.
Cryo-EM structures of the SARS-CoV-2 S trimer in complex with Fabs. All the structures were generated by rigid-body docking of the coordinates of S trimer subunits (PDB: 6vsb). The Fab-RBD-complexes reconstructed into the cryo-EM density maps and shown from both the side view (up) and top view (down). (A) Structures of S trimer in complex with A5–10 have three distinctive conformations. Left: only one RBD rotating up but each RBD binding with a Fab 5–10. Middle: two RBDs (one up and one down) binding with two Fabs and one RBD free. Right: three RBDs in ‘all-closed’ with only one accessible to Fab 5–10. (B) Structures of S trimer in complex with A34–2 revealed two conformations—one with RBDs in the ‘all-up’ configurations, each being recognized by a Fab (left); another with one RBD in down and two RBDs in up and only the ones in up binding with Fab 34–2.