Figure 4. Application of continuous evolution for studying drug resistance.
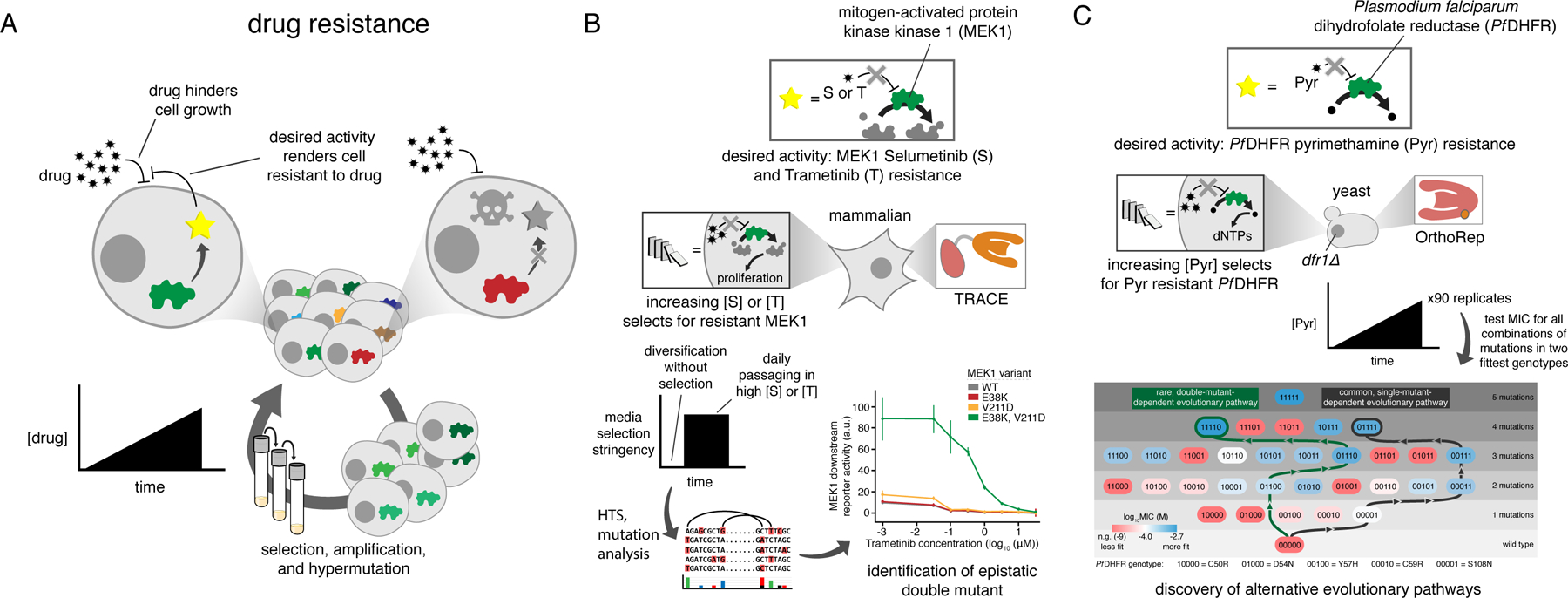
A) Generalized schematic for drug resistance continuous evolution experiments. A cycle with only one step is shown but is meant to represent continuous growth following desired selection schedules, for example serial passaging into media with increasing drug concentration. B) Continuous evolution of MEK1 with the TRACE MutaT7 system. The native activity of MEK1 is essential for proliferation in the mammalian cell line that was used (HEK293T) and is therefore drug selectable. MEK1 was targeted for mutagenesis and grown for a short initial period without selection for drug resistance to generate mutational diversity. Selection was then applied using a static high concentration of either of the two drugs being studied. C) Continuous evolution of DHFR to study drug resistance using OrthoRep. DHFR activity is essential for nucleotide biosynthesis and is therefore selectable in cells deleted for the native enzyme. Ninety replicate cultures were passaged in parallel with gradually increasing drug concentration, and Sanger sequencing of populations revealed two genotypes that commonly become fixed in populations. All combinations of the mutations in these two genotypes were individually cloned and tested in isolation to determine fitness and understand the accessibility of different mutational pathways leading to different outcomes. MIC, minimum inhibitory concentration.
