Figure 8.
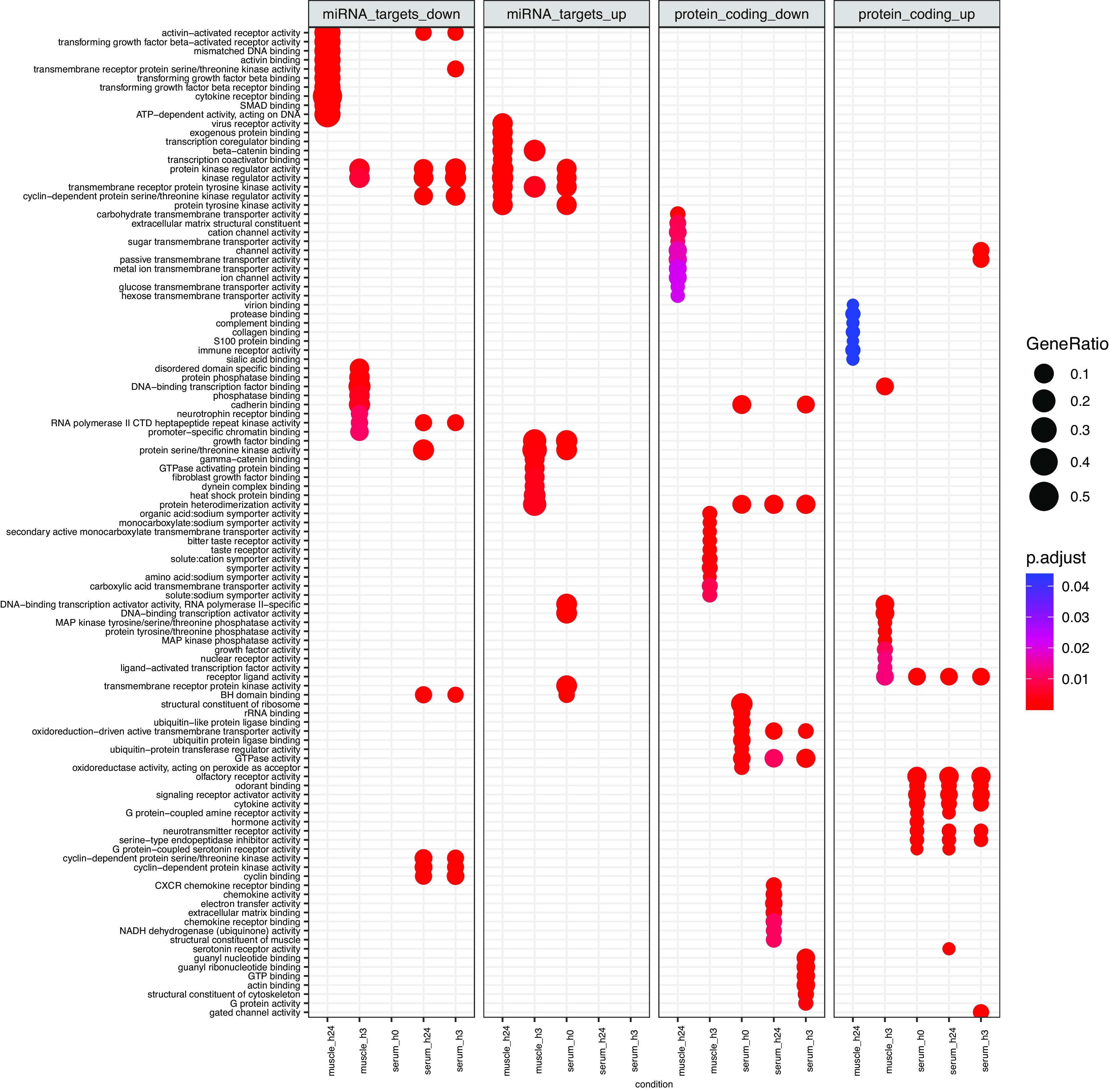
Biological processes associated with differentially expressed transcripts in skeletal muscle and serum extracellular vesicles at all postexercise time points (h0, h3, and h24) with false discovery rate < 0.05 and absolute log2fold-change >1 vs. preexercise, as curated by Gene Ontology. For miRNA targets (left two panels), multiMiR (v1.16) was used to identify the top 5% of predicted protein-coding mRNAs that functionally interact with miRNA differentially expressed following acute exercise. Targets were combined for all miRNA upregulated (up) or downregulated (down) at a given time point within muscle and serum extracellular vesicles and entered into Gene Ontology analysis. Differentially expressed protein-coding transcripts (right two panels) were directly input into Gene Ontology. GeneRatio (circle size) represents the proportion of genes shared between gene query and biological pathway, whereas likelihood of overlap based on the reference genome is represented by p.adjust (circle color).
