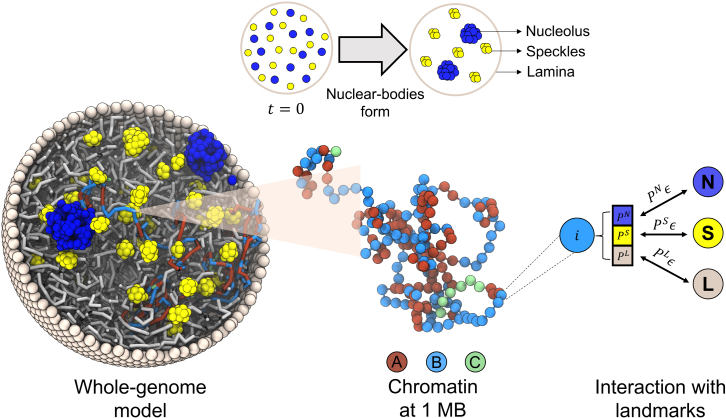
Figure 1.
Illustration of the whole-genome model that explicitly considers chromosomes and various nuclear landmarks, including lamina (gray), nucleoli (blue), and speckles (yellow). Chromosomes are modeled as beads-on-a-string polymers at 1 MB resolution where the beads are further classified into euchromatin (red, compartment A), heterochromatin (light blue, compartment B), and centromeric regions (green, compartment C). As shown by the schematic in the top panel, nucleoli and speckles form through the self-assembly of coarse-grained particles uniformly distributed inside the nucleus at the beginning of simulations. Coupling between chromosomes and nuclear landmarks is accounted for with specific interactions, the strength of which depends on the contact probabilities (, and ) between them quantified using high-throughput sequencing data (see text). To see this figure in color, go online.