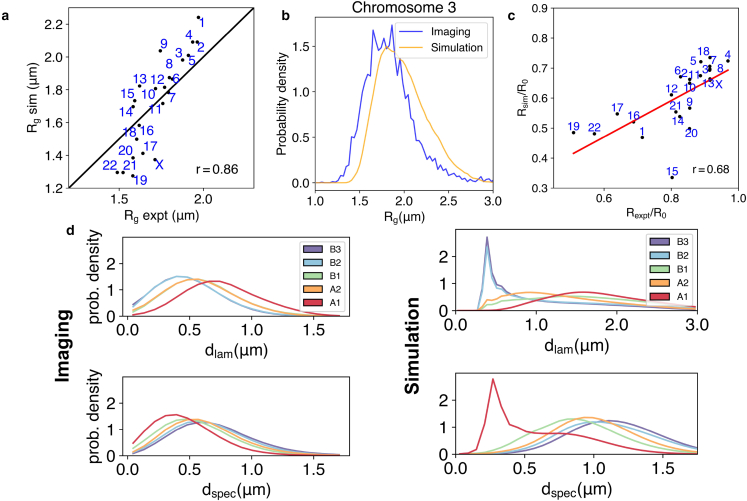
Figure 4.
Simulated structures reproduce chromosome sizes, positions, and the localization of genome subcompartments. (a) Comparison between chromosome radius of gyrations from simulations and from DNA-MERFISH experiments (28). The Pearson’s correlation coefficient () is also shown. (b) Overlap of the probability distributions for the radius of gyration of chromosome 3 computed from the experimental and simulated structural ensemble. (c) Comparison of the chromosome radial positions in experiment (88) and simulations. Here is the radius of the nucleus, and m. (d) Distribution of genome subcompartments as a function of distance from nuclear lamina () and speckles (), respectively, shown for DNA-MERFISH imaging (28) and simulation data. To see this figure in color, go online.