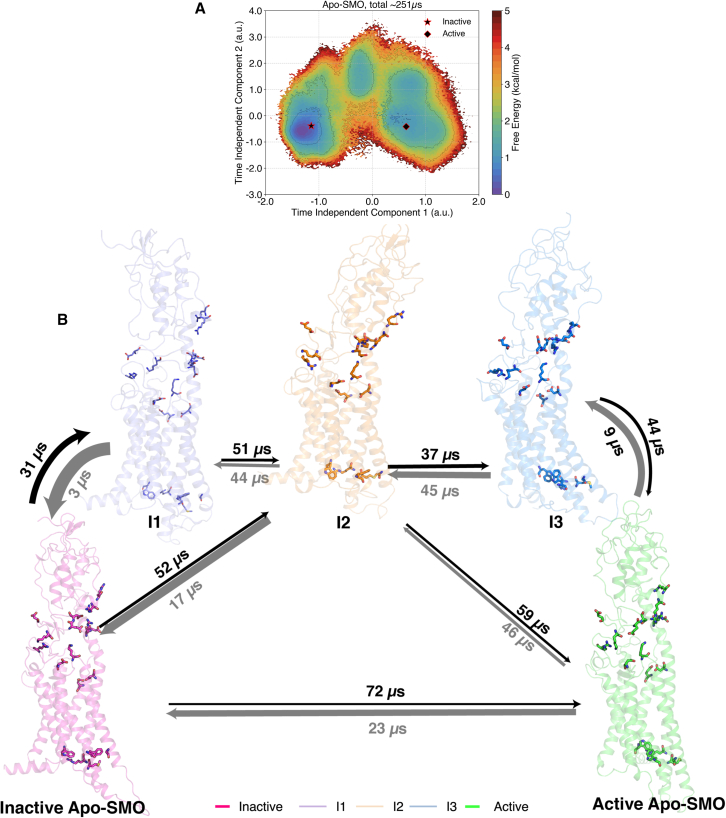
Figure 4.
(A) Relative free energies from MSM-weighted simulation data of Apo-SMO plotted along tIC1 and tIC2, the two slowest components, with the intermediate states as shown. The intermediate states were defined based on metastable basins and free-energy barriers associated with transitioning from an inactive to an active state. A cutoff of 1.8 kcal/mol was used to separate one basin from another. Residues shown as sticks include the π-cation lock, the WGM motif, and the salt bridges involved in activation. (B) Overall transition pathway of SMO activation process. The inactive (PDB: 5L7D) (32) and active (PDB: 6XBL) (37) structures are separated by the presence of three metastable conformations in between, . Residues shown by sticks correspond to the salt bridges, the WGM motif, the D-R-E network, and the π-cation lock, all residues critical for mediating SMO activation. To see this figure in color, go online.