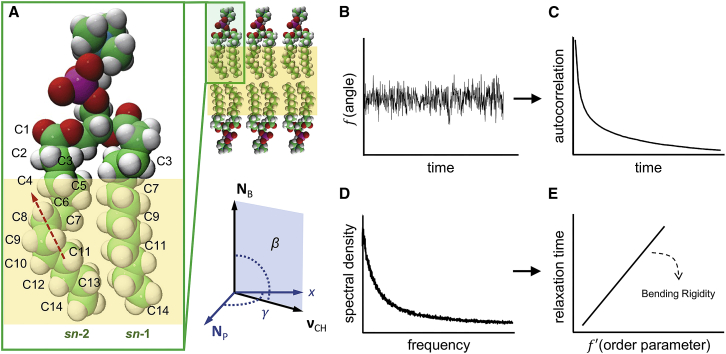
Figure 1.
Schematic representation of the simulation methodology. (A) Lipid bilayer and structure of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). Labels indicate carbons C3, C7, C9, C11, and C14 on the sn-1 chain and all carbons on the sn-2 chain of the lipid. Carbon atoms are shown in green, hydrogen in white, oxygen in red, and phosphorus in purple. Red dashed arrow shows a representative local director vector spanning carbons C8 and C11 on the sn-2 chain. Yellow shading outlines the part of the lipid chains (in the absence of cholesterol) where the fluctuations of the CH bonds follow a square-law dependence. Shown also is an example of the angle between a CH bond vector on the lipid chain and the bilayer normal (director axis), as well as the angle describing rotation of around . (B) Fluctuations of and over time are analyzed to obtain (C) their autocorrelation function averaged across all lipids and time. Fourier transformation of the autocorrelation function gives (D) the spectral density from which (E) the NMR relaxation rate is obtained. Multiple carbon atoms on the lipid chains are used together with their respective order parameters to calculate the bilayer bending modulus. See text for details. To see this figure in color, go online.