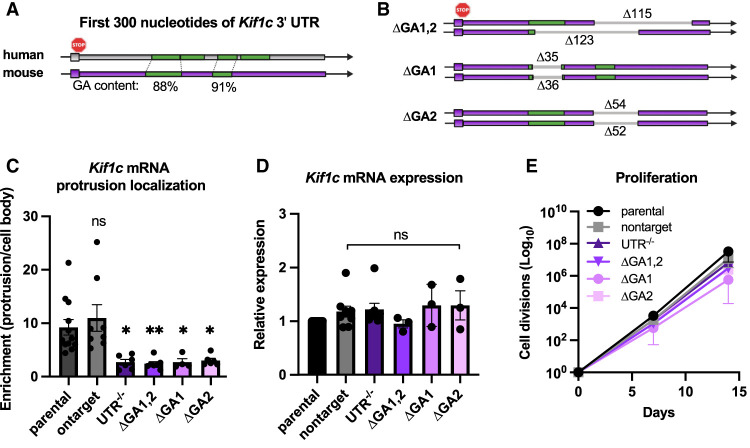
Figure 3.
The Kif1c mRNA localization element is made up of multiple GA-rich sequences. (A,B) Schematics of the first 300 nt of the Kif1c 3′ UTR. GA-rich elements (green) are highly conserved between mice and humans. (B) Schematic of endogenous deletions generated in this study. Unique deletions on each allele of the various ΔGA clonal cell lines are shown. (C,D) qRT-PCR of Kif1c mRNA in cells with endogenous deletions. All qRT-PCR data were normalized to Ppia and Ywhaz, which were validated to be stably expressed and uniformly distributed in all genetic contexts under study. All statistical tests are ordinary one-way ANOVA compared with parental. N = 3–9 biological replicates per condition. (**) P < 0.01, (*) P < 0.05. (E) Cell divisions measured every 2–3 d for 2 wk. N = 2–6 biological replicates per condition. Mean + SEM is shown for C–E.