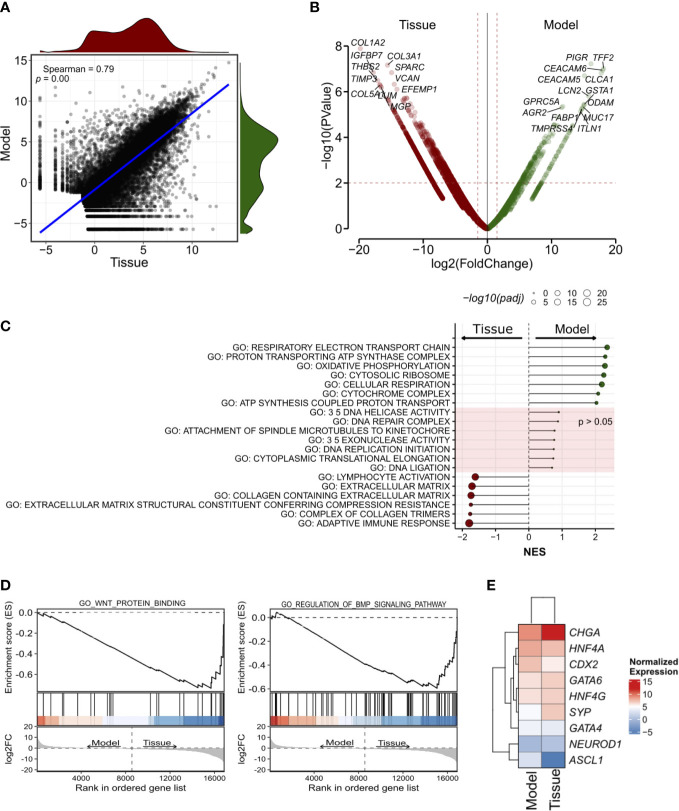
Figure 3.
Transcriptomic characterization of the siNET organoid culture. (A) Scatter plot showing the correlation between the model and the tissue transcriptomes (Spearman’s correlation) across all 19,828 genes detected. The density plots show the distribution of gene expression in the siNET organoid (green) and the tissue (red). (B) Volcano plot of differences in gene expression between tissue and model. Indicated are some of the genes with |log2FoldChange| ≥ 2 and adjusted p < 0.01. See Supplementary Table S3 for the complete list of differentially-expressed genes (DEGs). To account for the lack of biological and technical replicates in the identification of DEGs, we used a larger matrix of expression profiles of pancreatic ductal adenocarcinoma tissues and corresponding organoids. We used edgeR pipeline (27) to calculate a biological coefficient of variation (BCV) that ideally would represent an approximate biological variation for the siNET case. Squaring the BCV returned a dispersion value of 0.579 that was employed in the exactTest function to obtain p-values and fold-changes for each gene. (C) Selected GO pathways enriched in Model (green) or Tissue (red), or equally represented in both samples. GSEA was performed using gene sets from MsigDB library. See Supplementary Table 4 for the complete list of tested pathways. (D) GSEA plot showing enrichment of pathways involved in WNT and BMP signaling in patient’s tissue. (E) Heatmap showing the expression of transcription factors of the gastrointestinal cell lineage and neuroendocrine tumor markers in Tissue and Model.