ABSTRACT
Recently, long noncoding RNAs (lncRNAs) have been highlighted for extensive functionality in sepsis. In this study, we aimed to explore the role of RNF7 in the progression of sepsis. We initially established a rat model of sepsis through cecal ligation and puncture induction, whereupon RNF7 expression was determined by RT-qPCR. Following adenovirus infection, the role of RNF7 in muscle injury, skeletal muscle protein metabolism, oxidative stress, and inflammation in sepsis rats was analyzed. Then, downstream mechanisms of RNF7 were identified and validated. Further, lipopolysaccharide was applied to treat myoblast to further demonstrate the in vitro role of RNF7. Our results showed that RNF7 expression was upregulated during sepsis. Overexpression of RNF7 worsened the sepsis-induced skeletal muscle injury, induced skeletal muscle protein metabolism, oxidative stress, and inflammation in sepsis rats. Meanwhile, overexpression of RNF7 elevated thrombospondin-1 (THBS1) expression. Silencing of RNF7 inhibited THBS1 and activated the PI3K/Akt signaling pathway, arresting the release of inflammatory factors and oxidative stress levels in skeletal muscle cells. Altogether, RNF7 may promote skeletal muscle cell apoptosis while simultaneously inhibiting cell autophagy through the promotion of THBS1 and inactivation of the PI3K/Akt signaling pathway.
KEYWORDS: sepsis, long noncoding RNA RNF7, THBS1, PI3K/Akt signaling pathway, skeletal muscle cells, apoptosis, autophagy
INTRODUCTION
Sepsis is a complex and severe disorder that tends to induce systemic inflammation, immune suppression, and shock, exacerbating multiorgan failure. The incidence of sepsis has been sharply rising by 6% annually, and with a mortality rate of 26% (1). Accumulating evidence has elicited high short-term mortality for the disease; and sepsis additionally increases the risk of increased long-term mortality in survivors with sepsis, not only in developing countries, but also in developed countries (2). Fortunately, notable advancement in the therapeutic protocols has significantly reduced mortality; however, the prognosis remains poor due to the arrested development of improved treatments (3). Therefore, there is an urgent need for a comprehensive understanding of the disease pathogenesis and for the identification of effective therapeutic strategies.
Long noncoding RNAs (lncRNAs) are a diverse group of RNA sequences that serve as regulators of proteins and enzymes and interact with chromatin-modifying complexes, thereby extensively mediating several biological processes such as the immune response (4). Notably, lncRNAs can modulate numerous cancer-signaling pathways by interacting with proteins, RNA, and lipids and by facilitating or inhibiting the inflammatory process and the apoptosis of cancer cells (5). Several lncRNAs are implicated in sepsis based on their differential expression (6). For instance, lncRNA-CCL2 promotes inflammatory cytokine expression by interaction with the regulatory proteins in sepsis (7). Currently, only limited literature has identified the role of RNF7 in sepsis, although it has been identified that lncRNA RNF7 can exacerbate cardiac fibrosis in a rat sepsis model via thrombospondin-1 (THBS1) (8). THBS1 is essential for the regulation of both physiological and pathological skeletal muscle angiogenesis, a key process for normal skeletal muscle function (9). An existing study identified that the skeletal muscles (approximately 50% of body weight) are affected during acute and late sepsis, which is one of the sepsis-associated organ dysfunctions (10). Inhibition of skeletal muscle lactate release and atrophy exacerbate the suppression of sepsis progression (11). In light of the aforementioned literature, we speculated a potential detrimental role of RNF7 in inflammation during sepsis via THBS1. Sepsis can lead to severe and sustained muscle fiber atrophy in the limb muscles (12). Activation of phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) signaling pathway facilitates attenuation of sepsis-induced diaphragm atrophy (13). Moreover, RNF7 can activate the PI3K/Akt signaling pathway during cancer development, thus cooperating with protein regulation (14). In this study, we experimentally validated speculation that RNF7 may manipulate THBS1 expression and the PI3K/Akt signaling pathway, thus influencing the function of skeletal muscle cells during the development of sepsis in vivo and in vitro.
RESULTS
Successful establishment of cecal ligation and puncture (CLP) rat models.
To study the associated mechanism responsible for skeletal muscle cell injury, apoptosis, and autophagy in septic rats, we established a rat sepsis model. At 12 h after operation, the CLP-induced rats began to die, and the number of CLP-induced rat deaths continued to increase over time until it gradually stabilized. Among the rats, all 15 sham-operated rats survived, while 30 rats died in the postoperative period, and 90 CLP-induced rats survived (Fig. S1A). CLP-induced rats featured symptoms such as vertical hair, appetite loss, movement reduction, evident fatigue, and canthus secretions. In comparison with the sham-operated rats, the CLP-induced rats exhibited significant weight loss after a week (Fig. S1B). The preceding results are indicative of successful establishment of a rat sepsis model.
RNF7 promotes sepsis-induced skeletal muscle injury.
Existing literature has identified a high expression pattern of RNF7 in sepsis (8). Analysis of the GSE28750 data set revealed a notably high expression pattern of RNF7 in the sepsis samples (Fig. 1A). The results of fluorescence in situ hybridization (FISH) assay elicited predominant localization of RNF7 in the cytoplasm of skeletal muscle cells (Fig. 1B).
FIG 1.
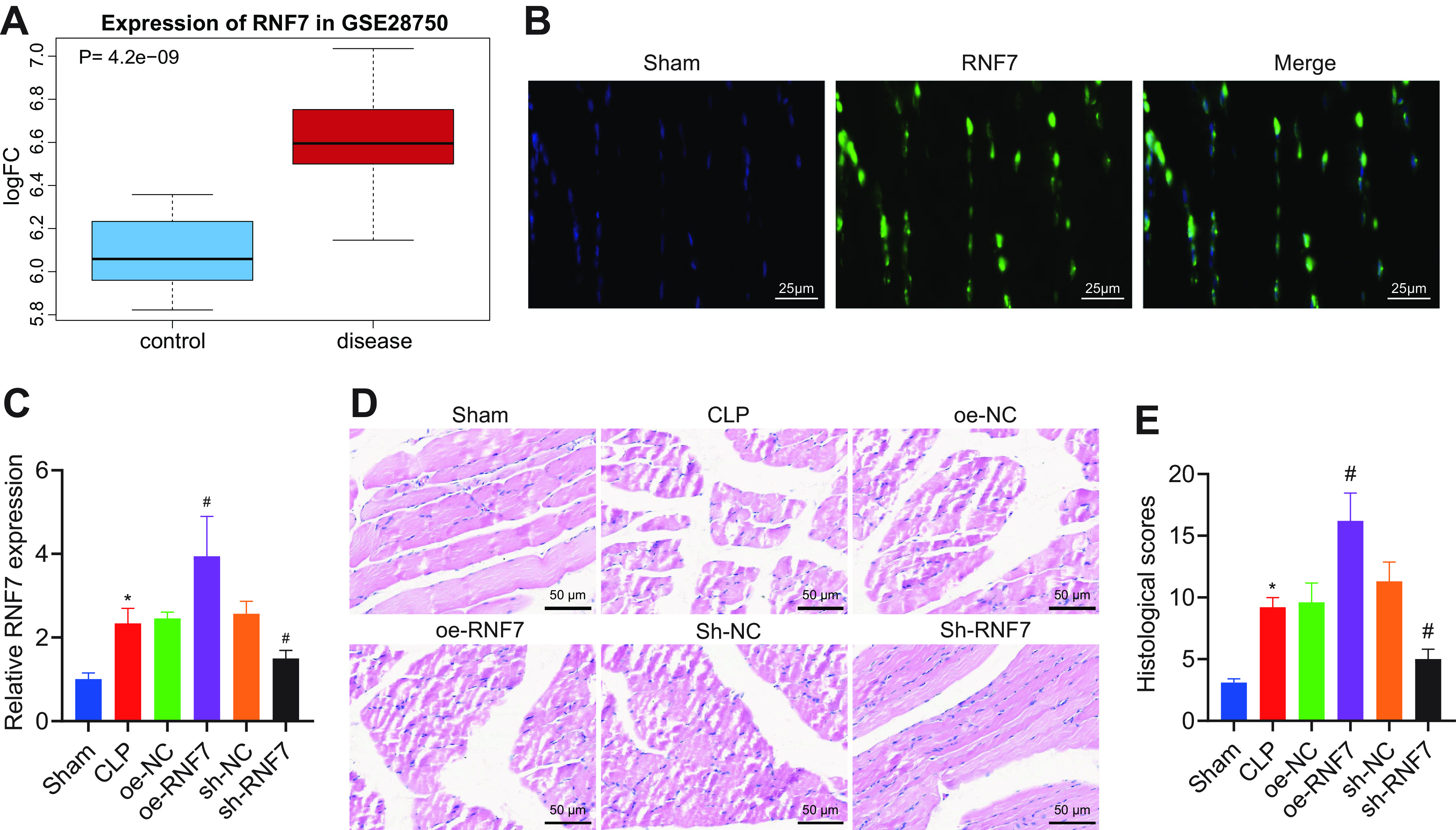
RNF7 is highly expressed in skeletal muscle tissues of septic rats and elicits sepsis-induced skeletal muscle injury. (A) Box plot of RNF7 expression pattern in sepsis samples in the GSE28750 data set, with blue box on the left indicating normal samples and red box on the right indicating sepsis samples. (B) Subcellular localization of RNF7 determined by FISH assay in skeletal muscle cells. (C) RT-qPCR analysis of RNF7 expression pattern in skeletal muscle tissues of CLP rats treated with oe-RNF7 or sh-RNF7. (D) Representative images of HE staining of skeletal muscle tissues of CLP rats treated with oe-RNF7 or sh-RNF7. (E) Pathological score of skeletal muscle tissues of CLP rats treated with oe-RNF7 or sh-RNF7. *, P < 0.05 versus sham-operated rats; #, P < 0.05 versus CLP rats treated with oe-NC or sh-NC. n = 10 for rats upon each treatment.
To study the effect of RNF7 on skeletal muscle damage in sepsis rats, sepsis rats were divided into two groups and then treated with overexpressed RNF7 (oe-RNF7) or short hairpin RNA (sh)-RNF7 (sh-RNF7), followed by CLP model establishment and reverse transcription quantitative PCR (RT-qPCR) detection. The results presented with an elevated expression pattern of RNF7 in the CLP-induced rats relative to the sham-operated rats. Additionally, an elevated RNF7 expression pattern was observed due to an overexpression of RNF7, while it was reduced following sh-RNF7 (Fig. 1C). Moreover, the rat liver, kidney, and lung tissues were harvested to detect the RNF7 expression pattern. No significant difference was found in the RNF7 expression pattern in the rat liver, kidney, and lung tissues upon individual treatment (Fig. S2). The preceding results indicate that overexpression or silencing of RNF7 in the rat skeletal muscle is specific and exclusive to the skeletal muscle.
According to hematoxylin-eosin (HE) analysis of the skeletal muscle tissues (Fig. 1D, E), the size of skeletal muscle of the sham-operated rats was normal, with darker color and no obvious pathological changes. In skeletal muscle tissues of CLP-induced rats, widening of the muscle space was identified with segmental necrosis of skeletal muscle cells, exudation of interstitial inflammatory cells, and increased pathological score of rat skeletal muscle tissues. Additionally, the oe-RNF7 treatment exhibited a more definitive increase in muscle space with segmental necrosis, exudation of interstitial inflammatory cells, and the pathological score of rat skeletal muscle tissues. Conversely, sh-RNF7 treatment elicited conflicting results relative to oe-RNF7 treatment.
The aforementioned results indicate that RNF7 is predominantly located in the cytoplasm of skeletal muscle cells, is highly expressed in the skeletal muscle tissues of septic rats, and can exacerbate sepsis-induced skeletal muscle injury.
RNF7 increases the skeletal muscle protein metabolism.
3-MH is a type of trace amino acid, principally present in skeletal muscles to reduce the contents of actin and myosin, and the measurement of 3-MH release can indirectly serve as an indicator for the degradation rate of myofibrillar protein (11). Analytical results of skeletal muscle protein metabolism revealed higher Tyr and 3-MH contents in the skeletal muscle tissues of CLP-induced rats relative to the sham-operated rats. Moreover, the overexpression of RNF7 elevated the Tyr and 3-MH contents, while silencing of RNF7 reduced their contents (Table S1). In summary, our results reveal that RNF7 can facilitate skeletal muscle protein metabolism in septic rats.
RNF7 promotes oxidative stress and inflammation in septic rats.
Next, we explored the underlying mechanism of RNF7 in respect to oxidative stress and inflammation in sepsis (Table S2, S3). In comparison to the sham-operated rats, the CLP rats exhibited elevated levels of malondihaldehyde (MDA) and NO with increased expression patterns of several inflammatory factors (TNF-α, IL-6, and IL-1β), as well as reduced activity of glutathione peroxidas (GSH-PX), superoxide dismutase (SOD), and chloramphenicol acetyltransferase (CAT). Treatment with oe-RNF7 resulted in elevations of NO and MDA as well as the levels of TNF-α, IL-6, and IL-1β; however, reductions were identified in the activity of GSH-Px and SOD, while silencing of RNF7 elicited conflicting results. Cumulatively, the preceding results elicited that RNF7 overexpression can induce oxidative stress and inflammation in septic rats.
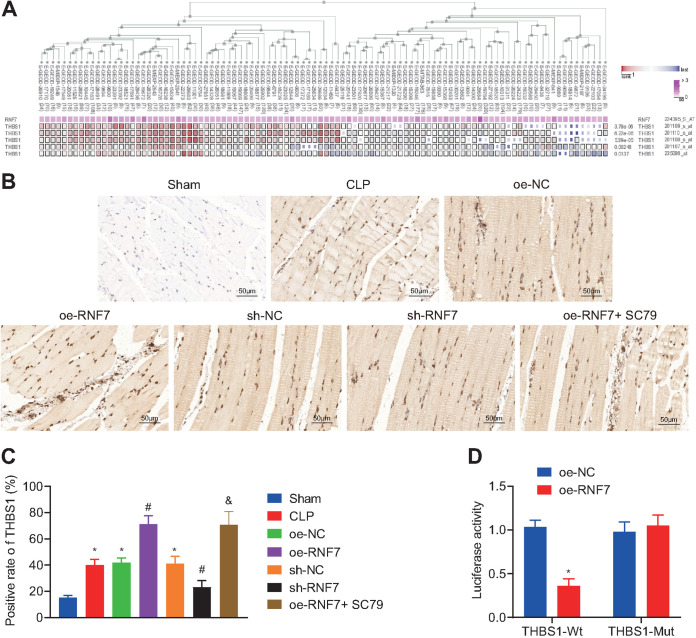
RNF7 upregulates THBS1 expression in septic rats.
The MEMW database predicted a coexpression of RNF7 with THBS1 (Fig. 2A). It has been reported that SC79 acts as a specific activator of the AKT signaling pathway (15, 16). Therefore, we then overexpressed RNF7 (oe-RNF7) in rats, then constructed a CLP model and performed SC79 treatment. The immunohistochemical staining results (Fig. 2B, C) revealed that THBS1-positive cells were brown-yellow particles with predominant localization of the THBS1-positive expression pattern in the cytoplasm and extracellular matrix. In comparison with the sham-operated rats, the THBS1 positive expression pattern was higher in the skeletal muscle tissues of the CLP rats. Additionally, an elevated THBS1-positive expression pattern was identified in the skeletal muscle tissues of CLP rats treated with oe-RNF7, while it was reduced in the sh-RNF7-treated CLP rats. Relative to oe-RNF7 treatment, both oe-RNF7 and SC79 exerted no affect in THBS1 positive expression. Moreover, the results from a dual luciferase reporter gene assay revealed that the luciferase activity of THBS1-3′-UTR wild type (WT) was reduced after RNF7 overexpression, which failed to alter the luciferase activity of 3′-UTR mutant type (MUT) (Fig. 2D), thus suggesting a direct interaction between RNF7 and THBS1. Collectively, the preceding results elicited that RNF7 can increase the THBS1 expression in the skeletal muscle tissues of septic rats.
FIG 2.
RNF7 promotes THBS1 expression pattern in the skeletal muscle tissue of septic rats. (A) MEM analysis of coexpression of RNF7 and THBS1. (B) IHC staining analysis of THBS1 protein in skeletal muscle tissues of rats treated with oe-RNF7 or sh-RNF7. (The injection was performed 1 week before operation with the volume of adenovirus set as 10 μL of the viral titer [1 × 1010 PFU/mL]). (C) Quantitative analysis of panel B. (The injection was performed 1 week before operation with the volume of adenovirus set as 10 μL of the viral titer [1 × 1010 PFU/mL]). *, P < 0.05 versus sham-operated rats; #, P < 0.05 versus CLP rats treated with sh-NC or oe-NC. (D) Interaction between THBS1 and RNF7 verified by dual luciferase reporter gene assay in HEK293T cells. *, P < 0.05 versus HEK293T cells transfected with oe-NC. n = 10 for rats upon each treatment. The cell experiment was conducted in triplicate independently.
RNF7 suppresses skeletal muscle cell autophagy and stimulates skeletal muscle cell apoptosis by increasing THBS1 and inactivating the PI3K/Akt signaling pathway in vivo.
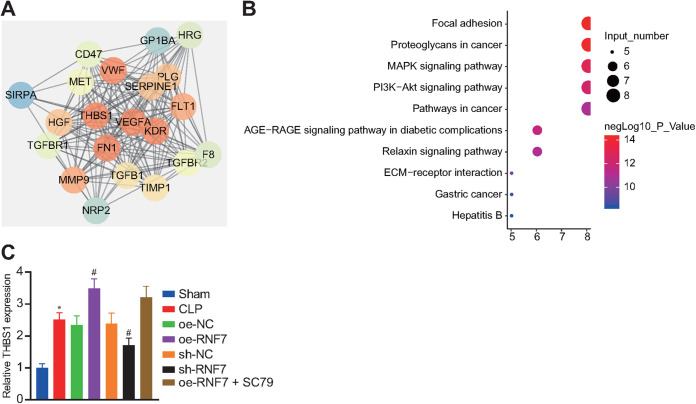
The STRING database predicted a total of 20 genes relative to THBS1 (Fig. 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of these 21 genes highlighted notable enrichment of 8 genes in the PI3K/Akt signaling pathway (Fig. 3B). The aforementioned results indicate that RNF7 may affect the PI3K/Akt signaling pathway by regulating THBS1 in sepsis. RT-qPCR detection results illustrated a higher THBS1 expression pattern in the skeletal muscle tissues of CLP rats compared to the sham-operated rats. An elevated THBS1 expression pattern was evident in the oe-RNF7-treated CLP rats, while conflicting results were apparent in the sh-RNF7 RNF7-treated CLP rats. No profound difference was identified in the THBS1 expression pattern between the oe-RNF7 + SC79 and the oe-RNF7-treated CLP rats (Fig. 3C).
FIG 3.
RNF7 augments THBS1 and inactivates the PI3K/Akt signaling pathway, thus stimulating skeletal muscle cell apoptosis and reducing cell autophagy. (A) PPI network of 20 genes related to THBS1 predicted by STRING. (B) KEGG enrichment analysis of these 20 genes. The ordinate refers to enriched signaling pathways, the abscissa refers to the number of genes enriched in the signaling pathway, and the bubble color indicates significance lg(p). (C) RT-qPCR analysis of THBS1 expression pattern in skeletal muscle tissues of CLP rats treated with oe-RNF7, sh-RNF7, or oe-RNF7 + SC79. *, P < 0.05 versus sham-operated rats; # P < 0.05 versus CLP rats treated with sh-NC or oe-NC; &, P < 0.05 versus CLP rats treated with oe-RNF7. n = 10 for rats upon each treatment.
Previous study has shown that Beclin-1 is one of two key proteins in autophagy, which can inhibit autophagy (17). Similarly, change of LC3-II/LC3-I ratio shows a significant positive correlation with autophagy, which is an effective indicator of autophagy (18). To investigate the effects of RNF7 on apoptosis and autophagy in skeletal muscle tissue of sepsis rats, we determined the levels of apoptosis-related proteins (Bax, Bcl-2) and autophagy-related proteins (Beclin-1 and LC3-II/LC3-I) by Western blotting. Western blot analysis suggested considerable elevations in THBS1 and Bax expression patterns, yet reductions were evident for Beclin-1, Bcl-2, and LC3-II/LC3-I, as well as PI3K and Akt phosphorylation levels in the skeletal muscle tissues of CLP rats relative to the sham-operated rats. Overexpression of RNF7 further elevated the THBS1 and Bax expression patterns, while reducing expression patterns of Beclin-1, Bcl-2, and LC3-II/LC3-I as well as PI3K and Akt phosphorylation levels. Opposing results were observed in the absence of RNF7. Additionally, expression patterns of Beclin-1, Bcl-2, LC3-II/LC3-I, p-PI3K/PI3K, and p-Akt/Akt were increased, while Bax was reduced upon both overexpression of RNF7 and SC79 relative to an individual overexpression of RNF7 (Fig. 4A, B).
FIG 4.
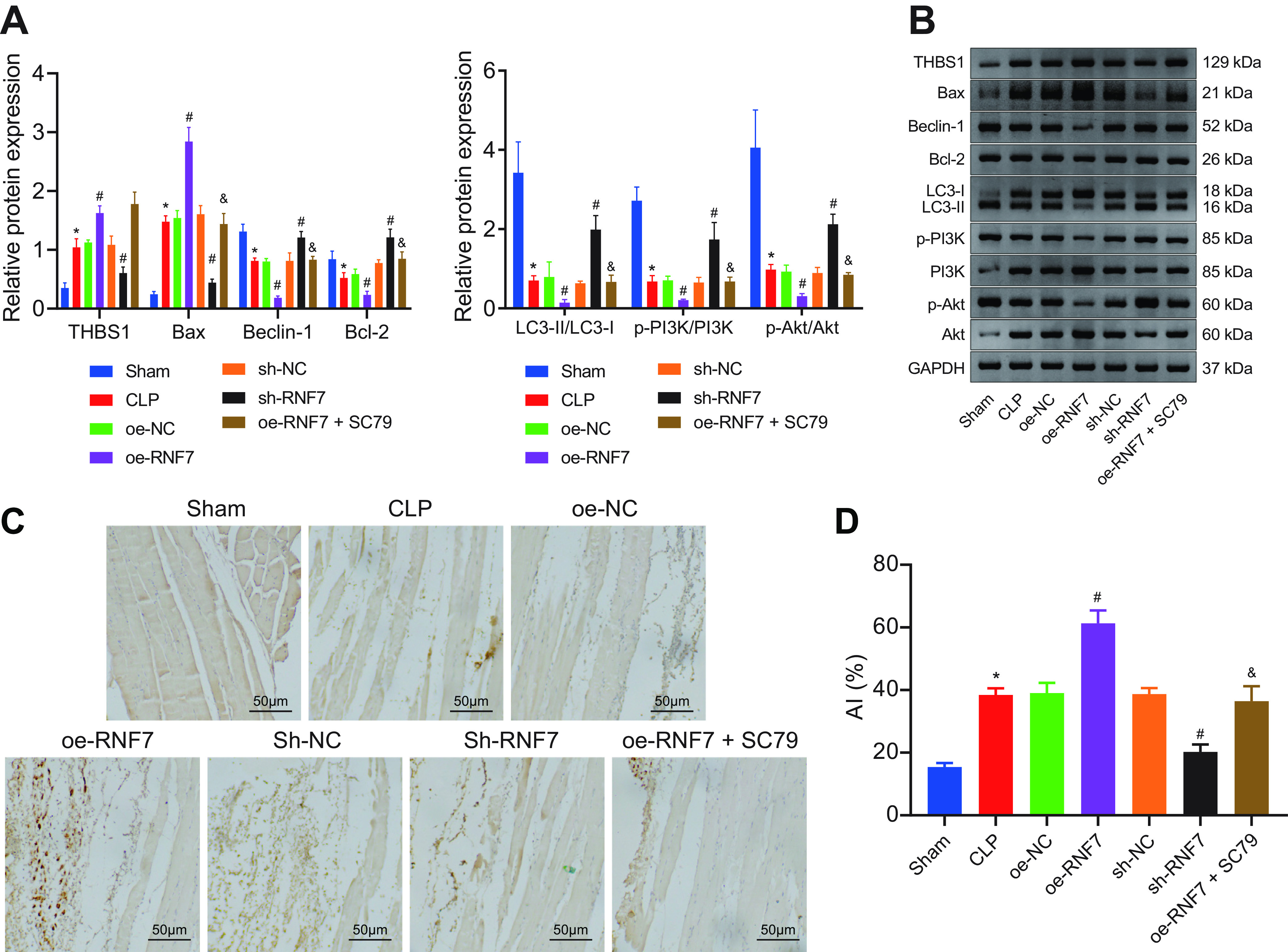
RNF7 suppresses skeletal muscle cell autophagy and stimulates the apoptosis of skeletal muscle cells through inactivation of the PI3K/Akt signaling pathway. (A and B) Analysis of expression of THBS1, Bax, Bcl-2, Beclin-1, and LC3-II/LC3-I proteins, as well as phosphorylation levels of PI3K and Akt in the skeletal muscle tissue of CLP rats treated with sh-RNF7, oe-RNF7, or oe-RNF7 + SC79 measured by Western blotting. (C and D). TUNNEL-positive cells in the skeletal muscle tissues of rats treated with oe-RNF7, sh-RNF7, or oe-RNF7 + SC79. *, P < 0.05 versus sham-operated rats, #, P < 0.05 versus CLP rats treated with sh-NC or oe-NC; &, P < 0.05 versus CLP rats treated with oe-RNF7. n = 10 for rats upon each treatment.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) staining data revealed a higher apoptosis rate of cells in the skeletal muscle tissues of CLP rats compared with the sham-operated rats. Moreover, oe-RNF7 treatment resulted in a significantly higher apoptosis rate, whereas sh-RNF7 treatment reduced the cell apoptosis rate. Treatment with oe-RNF7 + SC79 could annul the effect of oe-RNF7 (Fig. 4C, D). Conjointly, our results implicated that RNF7 promoted skeletal muscle cell apoptosis through inactivation of the PI3K/Akt signaling pathway.
Altogether, RNF7 can elevate THBS1 and inactivate the PI3K/Akt signaling pathway, thus inducing skeletal muscle cell apoptosis and reducing cell autophagy.
Silencing of RNF7 inhibits THBS1 and activates the PI3K/Akt signaling pathway, arresting the release of inflammatory factors and oxidative stress levels in skeletal muscle cells.
We then sought to explore the regulatory effect of RNF7 on myoblasts in vitro. RT-qPCR results demonstrated elevations in RNF7 and THBS1 expression patterns in lipopolysaccharide (LPS)-induced C2C12 cells relative to the control cells. However, LPS-induced cells treated with sh-RNF7 exhibited notable reductions in the RNF7 and THBS1 expression patterns. In the presence of RNF7 overexpression, the RNF7 and THBS1 expression patterns were elevated while further SC79 treatment led to no significant change (Fig. 5A). Additionally, Western blot analysis elicited higher THBS1 and Bax expression patterns along with lower expression patterns of Beclin-1, Bcl-2, and LC3-II/LC3-I as well as PI3K and Akt phosphorylation levels in the LPS-treated C2C12 cells relative to the control cells. In contrast, silencing of RNF7 resulted in opposite results. Additionally, RNF7 overexpression increased the THBS1 and Bax expression patterns, while reducing expression patterns of Beclin-1, Bcl-2, and LC3-II/LC3-I as well as the PI3K and Akt phosphorylation levels. Following subsequent SC79 treatment, the Bax expression pattern was reduced, while expression patterns of Beclin-1, Bcl-2, and LC3-II/LC3-I along with Akt phosphorylation level was elevated, along with unaltered PI3K phosphorylation level and THBS1 expression pattern (Fig. 5B). Moreover, enzyme-linked immunosorbent assay (ELISA) results demonstrated that NO, MDA, TNF-α, IL-6, and IL-1β expression patterns were increased, while the levels of GSH-Px and SOD along with CAT activity was decreased in LPS-treated C2C12 cells compared to the control cells. In contrast, silencing of RNF7 led to inhibition of NO, MDA, TNF-α, IL-6, and IL-1β expression patterns yet elevation in the levels of GSH-Px and SOD along with CAT activity. Besides, overexpression of RNF7 induced opposing results. A combination treatment protocol with oe-RNF7 and SC79 annulled the effect of an individual overexpression of RNF7 (Fig. 6A, B). Overall, the preceding results suggested that knockdown of RNF7 could downregulate THBS1 and activate the PI3K/Akt signaling pathway, thus preventing the release of inflammatory factors and oxidative stress levels in skeletal muscle cells.
FIG 5.
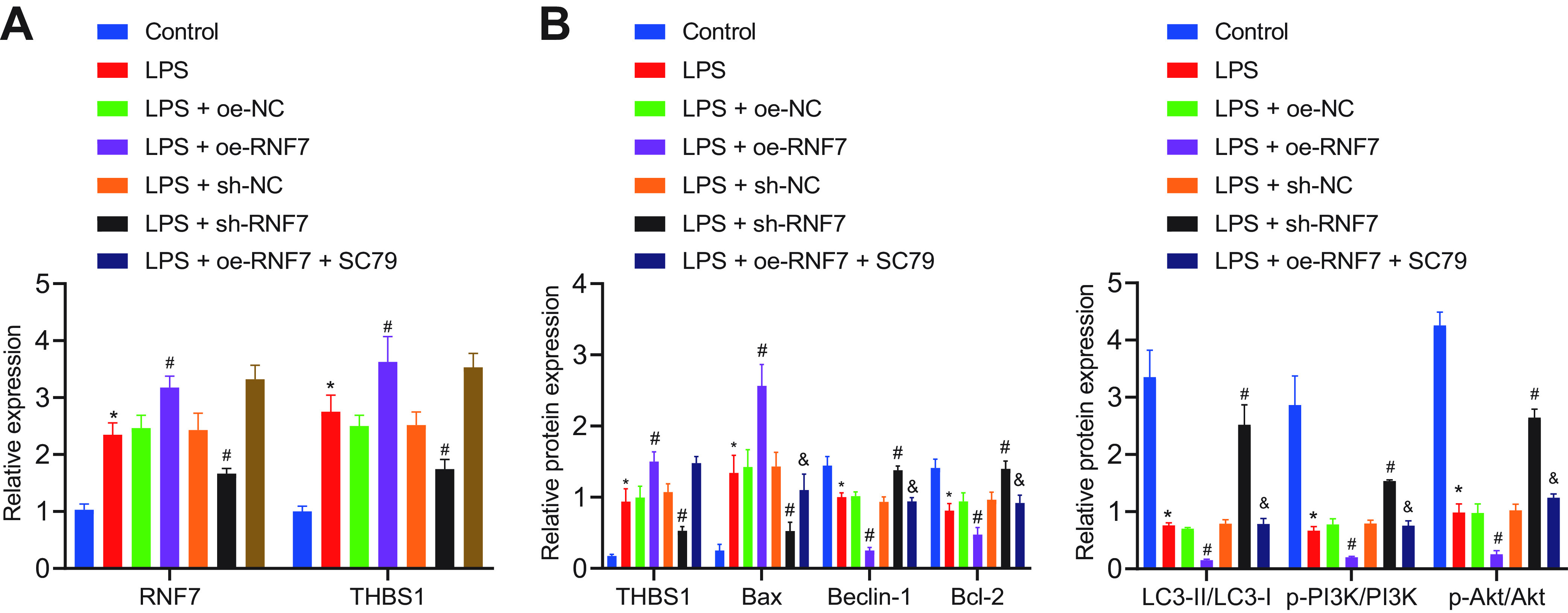
Knockdown of RNF7 decreases the release of inflammatory factors from the LPS-treated C2C12 cells and inhibits oxidative stress via the THBS1/PI3K/Akt axis in vitro. (A) Expression patterns of RNF7 and THBS1 in LPS-induced C2C12 cells treated with sh-RNF7, oe-RNF7, or oe-RNF7 + SC79 determined by RT-qPCR. (B) Analysis of THBS1, Bax, Bcl-2, Beclin-1, and LC3-II/LC3-I proteins, as well as phosphorylation levels of PI3K and Akt in LPS-induced C2C12 cells treated with sh-RNF7, oe-RNF7, or oe-RNF7 + SC79. *, P < 0.05 versus control cells; #, P < 0.05 versus LPS-treated cells; &, P < 0.05 versus LPS-induced cells treated with oe-RNF7. The cell experiment was carried out in triplicate independently.
FIG 6.
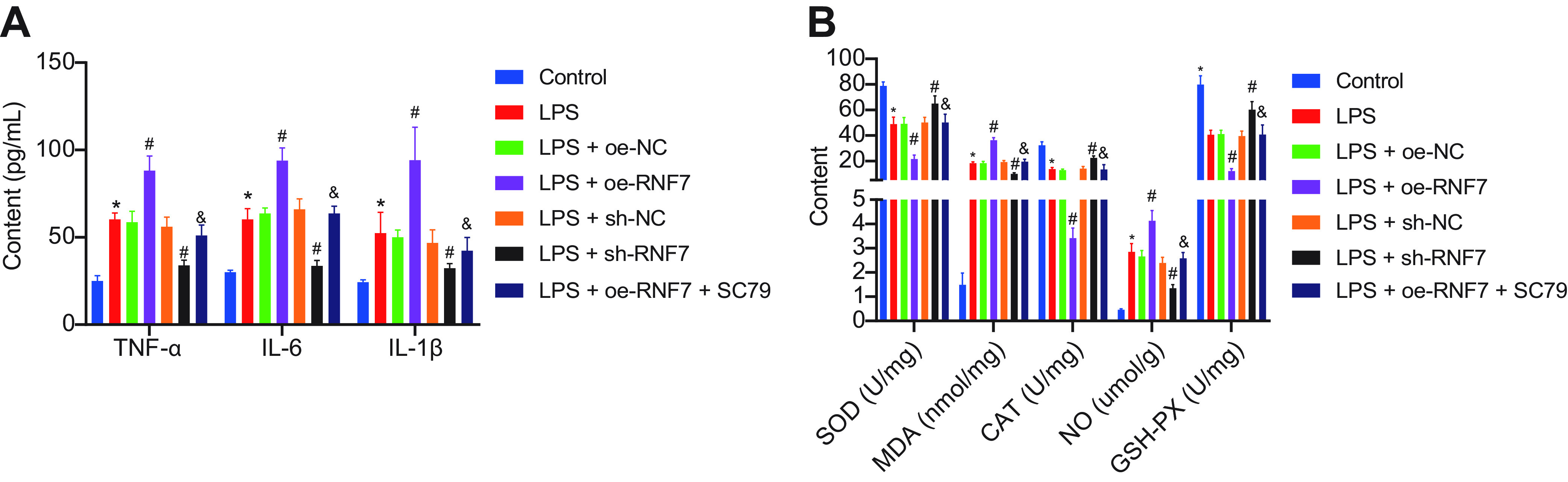
(A) Expression patterns of inflammatory factors measured by ELISA in LPS-induced C2C12 cells treated with sh-RNF7, oe-RNF7, or oe-RNF7 + SC79. (B) Expression patterns of oxidative stress-related factors measured by ELISA in LPS-induced C2C12 cells treated with sh-RNF7, oe-RNF7, or oe-RNF7 + SC79. *, P < 0.05 versus control cells; #, P < 0.05 versus LPS-treated cells; &, P < 0.05 versus LPS-induced cells treated with oe-RNF7. The cell experiment was carried out in triplicate independently.
DISCUSSION
Sepsis is a systemic inflammatory disease mediated by the innate immune system, which presents with deleterious complications such as kidney injury, skeletal muscle disorders, or shock (3). In this study, we established a CLP-induced sepsis rat model to determine the functionality of RNF7 in sepsis. Our results elicited that RNF7 increased the production of inflammatory factors as well as apoptosis and facilitated sepsis progression through the inactivation of the PI3K/Akt signaling pathway.
Initially, our findings demonstrated the upregulation of RNF7 in the skeletal muscle tissues of septic rats with severe skeletal muscle injury in sepsis. An existing study identified interactions of lncRNA with several biological molecules to regulate diverse cellular processes in various diseases (19), including sepsis. Accumulating studies have determined an inhibitory effect exerted by the downregulation of lncRNA NEAT1 on immunity in septic rats by inhibiting mast cell expression of membrane protein 1 (20). Moreover, lncRNA MALAT1 also exhibits the potential as a definitive predictor for disease severity, inflammation, and survival of patients with sepsis (21). Additionally, an existing study determined the ability of downregulation of lncRNA MALAT1 to prevent excessive autophagy and apoptosis via the PI3K/Akt signaling pathway (22).
Specifically, our findings identified that overexpression of RNF7 can increase the apoptosis of skeletal muscle cells via activation of the PI3K/Akt signaling pathway. RNF7 can terminally bind to conserved sites of immunoglobulin molecules to induce the apoptosis of immune cells, as illustrated by the increased Bax protein levels and decreased Bcl-2 levels (23). Bcl-2, the most primitive member of the Bcl-2 family of apoptosis regulatory proteins, functions as an anti-apoptotic gene (24). Bax is also a member of the Bcl-2 family and functions as a major proapoptotic protein for regulating apoptosis in normal and cancer cells (25). Accumulating evidence has validated the ability of several lncRNAs to suppress activation of the PI3K/Akt signaling pathway (26, 27), while the inhibitory effect of RNF7 on the PI3K/Akt signaling pathway warrants further investigation due to the lack of supporting literature. Moreover, the inhibition of the PI3K/Akt signaling pathway can dramatically facilitate the apoptosis of myoblasts (28). Additionally, activation of the PI3K/Akt signaling pathway reduces oxidative stress and inflammation after ischemia-reperfusion injury (29). As for sepsis, capsaicin pretreatment can inhibit oxidative stress by upregulating the levels of its markers SOD and GSH-Px while downregulating the MDA level to exercise a protective effect for cardiomyocytes against LPS-induced damage to augment autophagy (30). Additionally, an existing study verified the ability of apigenin treatment to improve sepsis-induced lung injury by inhibiting the degree of oxidative stress, as demonstrated by increased SOD levels, CAT activity, and GSH levels (31). Additionally, our ELISA results revealed that RNF7 overexpression increased the levels of TNF-α, IL-6, and IL-1β in rat skeletal muscle tissue and cells during sepsis. Notably, RNF7-overexpressing macrophages elicited upregulation of several protumorigenic cytokines (IL-1b, IL-6, and TNF-α) (32). Strikingly, RNF7-depleting neutrophils released abundant concentrations of proinflammatory cytokines and with minor concentration of macrophages, while the depletion of RNF7 impeded the inflammatory responses in sepsis (33).
Another vital finding from our study elicited that RNF7 promoted autophagy in skeletal muscles during sepsis, as evidenced by the markedly elevated Beclin-1 and LC3 levels. Indeed, autophagy is a process for lysosomal degradation of cellular debris, consequently generating energy and macromolecular precursors, thus eliciting an ideal target for cancer treatment (34). The autophagy components determine the regulation of infectious agents while simultaneously hindering inflammatory pathology (35). Essentially, in sepsis, skeletal muscles are increasingly prone to sepsis-induced mitochondrial injury, decreased biogenesis, and increased autophagy (36). However, enhanced autophagy can serve as a protective barrier against multiple organ injuries in murine sepsis models, thereby preventing apoptosis and sustaining a homeostatic balance between the pro- and anti-inflammatory cytokines (37). Further studies are needed to explore the interaction between increasing apoptosis and autophagy in sepsis.
Fundamentally, our results identified an explicit association between the stimulative effect of RNF7 on cell apoptosis, inflammation, and oxidative stress, as well as the inhibiting effect on cell autophagy with THBS1 overexpression. In part consistent with this result, lncRNA NEAT1 exacerbated inflammation and apoptosis through THBS1 upregulation (38). THBS1 overexpression resulted in increased peritoneal inflammation and mortality, whereas THBS1 deficiency elicited a protective effect in septic rats (39). THBS1 can regulate the adhesion and migratory capacity of monocytic cells, inducing aortic expansion (40). In addition, activation of the PI3K/Akt signaling pathway is associated with the downregulation of THBS1 in vascular endothelial cells of rats with diabetic retinopathy (41). Consistently, the current study demonstrated that RNF7 promoted THBS1 through the PI3K/Akt signaling pathway inactivation and functioned in skeletal muscles during sepsis.
To conclude, RNF7 could promote skeletal muscle cell apoptosis and inhibit cell autophagy in sepsis by upregulating THBS1 and inactivating the PI3K/Akt signaling pathway (Fig. 7). This finding may contribute to the development of novel treatment protocols for sepsis, although further studies are required to validate the role of RNF7 in other tissues in sepsis.
FIG 7.
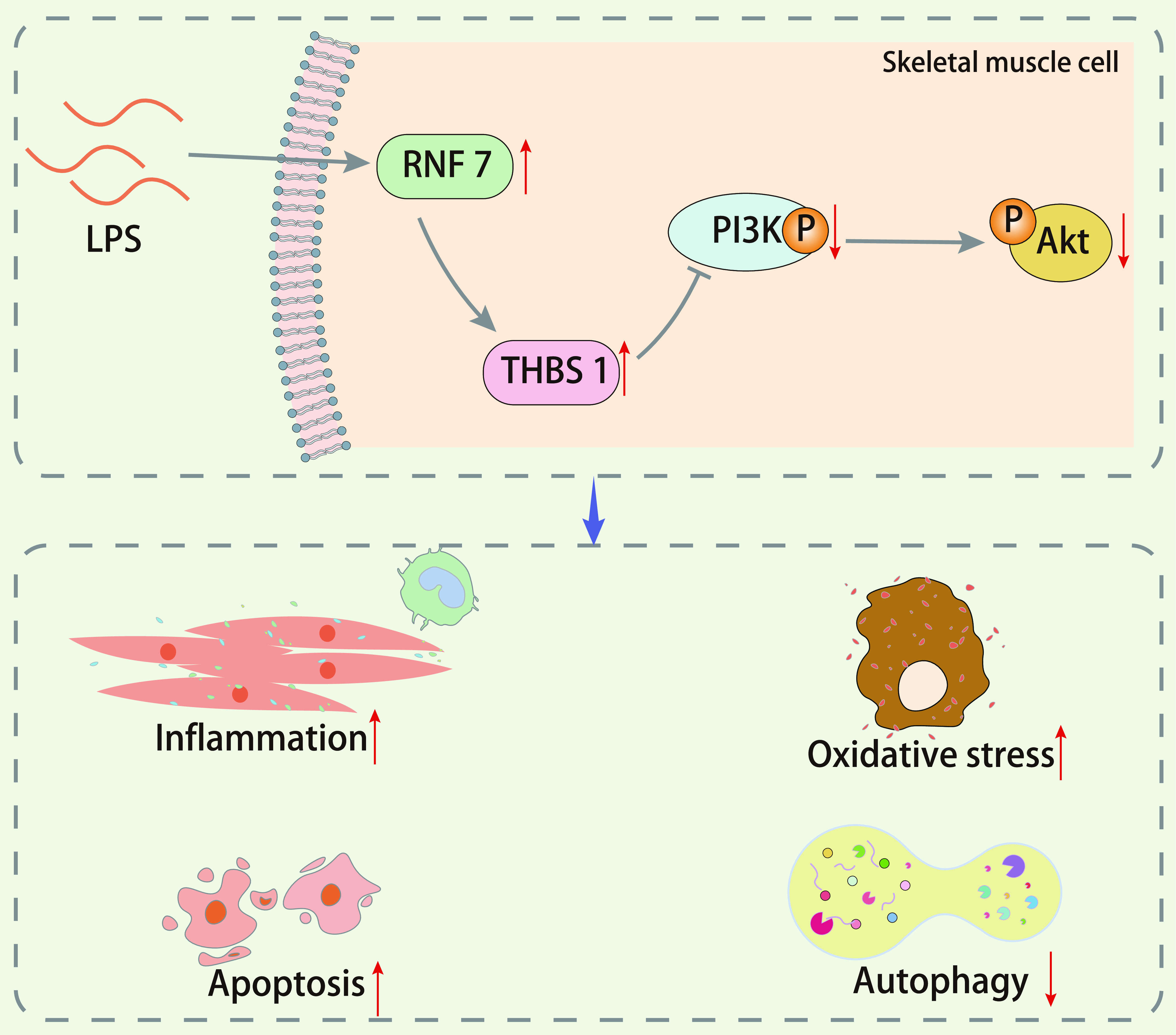
Schematic diagram of the mechanism by which RNF7 affects skeletal muscle cell apoptosis and autophagy in a rat sepsis model. RNF7 inhibits the PI3K/Akt signaling pathway by promoting the expression pattern of THBS1, thereby promoting the apoptosis of skeletal muscle cells and inhibiting their autophagy in a rat sepsis model.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed with approval of the Animal Ethics Committee of the First Hospital of Jilin University. The feeding of all animals in the experiment and the experimental operation was in compliance with the Regulations for the Administration of Affairs Concerning Experimental Animals.
Microarray-based gene expression profiling.
The regulatory pathways involved in the manifestation of sepsis were predicted through existing literature. The GSE28750 data set was retrieved from the Gene Expression Omnibus database to verify expression of several lncRNAs. Coexpression analysis was conducted by MEM to verify the relationship between lncRNAs and their downstream factors. SRING was applied to predict the relative genes of downstream genes. A protein–protein interaction network was constructed using Cytoscape. KOBAS was adopted to perform KEGG enrichment analysis to determine the enriched signaling pathway.
Establishment of CLP-induced rat sepsis models.
Male Sprague-Dawley rats (aged 12 to 15 weeks, weighing 202 ± 20 g; 101, Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were housed in a pathogen-free laboratory at 18 to 25°C with free access to chow and water. A sepsis model was established via CLP as per a previous protocol under 2% pentobarbital anesthesia (50 mg/kg body weight). The abdomen was fixed upward on the polyethylene foam board, and the middle and lower abdomen were selected for preparation of the skin. The skin was disinfected using iodophor, covered with disposable sterile gauze, and incised (length 3 to 4 cm) longitudinally along the midline of the abdomen. The cecum was extracted using a pair of toothless forceps, without damaging the mesangial blood vessels. The contents in the cecum were gently pushed to the distal end of the cecum to ensure that no obvious gas was present in the ligated cecum section, and the RM mesangium was separated. The dismal cecum was labeled with methylene blue dye (Shanghai Regal Biology Technology Co., Ltd., Shanghai, China) where blue label was evident in areas surrounding the cecum. The length of ligation was 1.5 cm, and the corresponding volume was 2.5 mL. Since the ligation positions were lower than the interstitial valve position, intestinal obstruction would not occur. The cecum was punctured using a 21-gauge needle from the mesenteric side to the opposite side following the single penetrating protocol. After the puncture, a small amount of feces at the two puncture holes were squeezed to promise that the perforation was open and functional. After ligation, the muscle layer and skin were sutured for complete closure of the abdominal cavity, after which prewarmed saline (37°C, 5 mL/100 g body weight) was subcutaneously injected for fluid replacement. Sham-operated rats that underwent only laparotomy without cecal ligation and perforation served as the negative-control (NC) group. All operations were expected to be completed within 10 min. After 2 days, rats were euthanized with pentobarbital at a dose of 100 mg/kg, after which the skeletal muscle, liver, kidney, and lung tissues were harvested for subsequent experiments.
Adenovirus vector construction and animal treatment.
Recombinant plasmids pShuttle2-EGFP-RNF7 and pShuttle2-EGFP-sh-RNF7 were transferred into the DH5a-competent cells for adenovirus packaging and amplification, with pShuttle2-EGFP serving as control. The pShuttle2-EGFP-RNF7, pShuttle2-EGFP-sh-RNF7, and pShuttle2-EGFP were digested by incubation with I-Ceu I and PI-Sce I to isolate the DNA fragments. The fragments were introduced into an adenovirus skeleton plasmid, pAdeno-X, using the Adeno-X Expression System kit (Clontech, Mountain View, CA, USA; K1650-1) to form oe-RNF7 or oe-NC. The recombinant adenovirus vector or the empty vector sh-RNF7 was transfected into the competent cells for amplification.
Sham-operated rats were used as control, and the CLP rats were further injected with adenovirus carrying oe-NC (model rats, skeletal muscle injected with adenovirus constructed with an overexpression vector without any sequence inserted), oe-RNF7 (model rats, skeletal muscle injected with RNF7 overexpressing adenovirus), sh-NC (model rat, skeletal muscle injected with adenovirus constructed with an interfering vector without any sequence inserted), or sh-RNF7 (model rats, skeletal muscle injected with interfering RNF7 adenovirus) or cotreated with oe-RNF7 + SC79 (an Akt activator, HY-18749, MedChemExpress Company, USA). The injection was performed 1 week before operation, with the volume of adenovirus set as 10 μL of the viral titer (1 × 1010 PFU/mL). SC79 was dissolved in DMSO (D2650, Sigma-Aldrich Chemical Company, St. Louis, MO, USA) at a concentration of 4 μg/mL. Ten rats were randomly selected from among the sham group, and 70 rats were randomly selected from the CLP group, for a total of 10 rats in each group. According to the sequence of RNF7 in the GenBank data set, the fragments and RNF7-related sequences were designed and synthesized by Shanghai Genechem Co., Ltd. (Shanghai, China).
HE staining.
Skeletal muscle tissues of rats were fixed using 4% paraformaldehyde for 24 h, dehydrated with gradient alcohol, and embedded in paraffin. The paraffin tissues were incised into 5-μm-thick sections, which were subjected to HE staining (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) (42). Finally, the samples were observed and documented under an optical microscope (XP-330, Shanghai Bingyu Optical Instrument, Co., Ltd., Shanghai, China).
Immunohistochemistry (IHC).
Paraffin-embedded tissues were incised into 4-μm-thick sections. Sections were dewaxed with xylene, rehydrated with gradient alcohol, and immersed in 3% H2O2 and subsequently in 4% formaldehyde (BD5024, Bioworld, USA) at 37°C for 30 min, followed by antigen retrieval with 0.01 M citric acid buffer. After a blockade with goat serum, the sections were probed with mouse anti-human THBS1 (at a dilution ratio of 1:500; ab85762, Abcam, Inc., Cambridge, UK) for 12 h. The following day, the sections were reprobed with biotinylated secondary antibody goat anti-mouse (A21020, Abbkine, USA) for 10 min and then treated with streptavidin-horseradish peroxidase (DF7852, Shanghai Yaoyun Biotechnology Co., Ltd., Shanghai, China) for 10 min. Finally, sections were developed using 3,3′-diaminobenzidine tetrahydrochloride and counterstained with hematoxylin. Using ImageJ analysis software (Eclipse 80i, Nikon, Japan), the positive cell ratio was calculated under a light microscope with five randomly selected fields: positive cells/all cells >10% (+), <10% (−).
Determination of proteolytic rate.
High-performance liquid chromatography was carried out to determine the content of 3-methylhistidine (3-MH) and tyrosine (Tyr) in muscle tissues as an existing protocol. Muscle tissue samples were incubated with the freshly oxygenated Krebs-Henseleit incubation medium with 0.5 mmol/l cycloheximide for 3 h to prevent the participation of the released amino acids in protein synthesis. The tissues were then suspended in perchloric acid (500 μL) and subjected to centrifugation at 17,510 × g for 25 min. For 3-MH determination, the supernatant or incubation medium was supplemented with a combination of sodium borate, acetonitrile, perchloric acid, and NaOH to maintain the pH at 6.0. Next, 3-MH was isolated from the mixed solution and was measured using UV spectroscopy at the excitation wavelength of 365 nm in sodium phosphate buffer. As for Tyr, the supernatant or incubation medium was reacted with OPA solution and analyzed with a Hypersil AA-ODS column (2.1 mm × 200 mm) the gradient elution method. Tyr content was measured at the excitation wavelength of 338 nm using an UV spectrophotometer (UV1901, Shanghai AuCy Instrument Co., Ltd., Shanghai, China).
Measurement of oxidative stress-related factors and inflammatory factors.
The levels of SOD (BC0170, Solarbio), MDA (A003-1, NanJing JianCheng Bioengineering Institute, Nanjing, China), TNF-α (JianCheng), GSH-PX (A005, Solarbio), blood urea nitrogen (BU7120, Beijing Lideman, Biotechnology Co., Ltd., Beijing, China), and Na+-K+-ATP enzyme (QS1700, Solarbio) in the supernatant of skeletal muscle tissues were detected in strict accordance with the corresponding kit instructions. Additionally, ELISA was performed to measure the levels of the inflammatory factors TNF-α, IL-6, and IL-1β in the supernatant of skeletal muscle tissues in strict accordance with the provided instructions (69-30484, 69-30490, and 69–30375, Mokesha Pharmaceutical Chemical Co., Ltd., Wuhan, China).
RT-qPCR.
The total RNA content was extracted from the skeletal muscle tissues of rats using the TRIzol reagent (Invitrogen, Carlsbad, Ca, USA). The RNA content was reverse-transcribed into cDNA with a PrimeScript RT kit (RR036A, TaKaRa, Japan). RT-qPCR was started on the basis of the instructions of the SYBR Premix Ex Taq II kit (TaKaRa, Japan) with the ABI PRISM 7300 system (Shanghai Kunke Instrument Co., Ltd., Shanghai, China). The transcriptional level of mRNA was estimated based on the 2-ΔΔCt quantification method, with β-actin serving as a normalizer. Primer sequences were designed by Bio Just Biomedical Science and Technology Co., Ltd. (Wuhan, Hubei, China), as shown in Table S4.
Western blot analysis.
The total protein content was extracted from the skeletal tissues using 1 mL of lysis buffer (50 mmol/L Tris, 150 mmol/L NaCl, 5 mmol/L EDTA, 0.1% SDS, 1% NP-40, 5 μg/mL Aprotinin, and 2 mmol/L PMSF), and the protein concentration was determined using a bicinchoninic acid kit (P0012, Shanghai Beyotime Biotechnology Co., Ltd., Shanghai, China). The protein was subject to separation by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PE0025, Beijing Leagene Biotech Co., Ltd., Beijing, China) and transferred onto nitrocellulose membranes. A membrane blockade was conducted using 5% skimmed milk powder for 1 h at room temperature, after which the membranes were probed at 4°C overnight with the primary antibodies and then reprobed with horseradish peroxidase-conjugated secondary antibody or IgG antibody (1:10,000, Jackson, USA) for 1 h. Next, the enhanced chemiluminescence solution (Pierce Biotechnology Inc., Rockford, IL, USA) was added for visualization of the X-ray results. With β-actin serving as internal reference, the relative protein expression was estimated as the ratio of the gray value of the target band to β-actin. The primary antibodies (Abcam) used in the preceding assay included THBS1 (ab85762), PI3K (1:2,000, ab180967), Akt (1:500, ab8805), phosphorylated (p)-Akt (1:750, ab38449), p-PI3K (1:1,000, ab191606), Bax (1:750, ab53154), Beclin-1 (1:1,000, ab207612), Bcl-2 (1:1,000, ab59348), light chain 3-II (LC3-II) (1:2,000, ab192890), and LC3-I (1:50,000, ab52628).
TUNEL staining.
TUNEL staining was performed to evaluate the degree of cell apoptosis. Briefly, the paraffin-embedded tissues were subdivided into 20-μm-thick sections. The sections were dewaxed with xylene, rehydrated with gradient alcohol, fixed with paraformaldehyde, and finally blocked with a combination of methanol and 3% H2O2. Subsequently, the sections were subjected to TUNEL staining in strict accordance with the provided instructions of the TUNEL kit (Roche Diagnostics GmbH, Mannheim, Germany) and then observed under optical microscopy (Leica DM4 P, Leica Biosystem, Shanghai, China) from 10 randomly selected visual fields to estimate the concentration of apoptotic cells. Finally, the apoptotic index (AI) of skeletal muscle cells was estimated as AI = apoptotic cells/total cells.
FISH.
Cy3-labeled RNF7 RNA-FISH was provided by Guangzhou RiboBio Co., Ltd. (Guangdong, China). FISH assay was performed in strict accordance with the provided instructions of the FISH kit (C002, Shanghai Gefan Biological Technology Co., Ltd., Shanghai, China).
Dual luciferase reporter gene assay.
The THBS1 3’ untranslated region dual luciferase reporter gene vector and mutant plasmids with mutations in the binding site of RNF7 were constructed: PmirGLO-THBS1-WT and PmirGLO-THBS1-MUT, respectively. The reporter plasmids were cotransfected with oe-RNF7 or its NC into HEK293T cells (ACS-4500, American Type Culture Collection [ATCC], Manassas, VA, USA). After transfection for 24 h, cells were lysed and centrifuged at 6,000 × g for 1 min, with isolation of the supernatant. The luciferase activity was detected using the Dual-Luciferase Reporter Assay System (E1910, Promega, Madison, WI, USA).
Culture and treatment of skeletal muscle cells.
Rat myoblast cell line (L6; CRL-1458, ATCC) was cultured with DMEM (Sigma-Aldrich) containing 20% FBS (PAN Biotech, Aidenbach, Germany) and 1% penicillin-streptomycin solution (Invitrogen) in a cell incubator with 5% CO2 (HeraCell, ThermoFisher Scientific, Rockford, IL, USA) at 37°C. Upon attaining 80 to 95% confluence, the cells were digested by the Trypsin-EDTA (Sigma-Aldrich) enzyme digestion method and isolated. The cells were seeded in a 12-well plate at a density of 12.5 × 103 cells/mL in each well. Approximately 3 days later, the medium was changed to the cell differentiation medium: DMEM + 2% horse serum (Sigma-Aldrich) with 1% penicillin-streptomycin solution. After 3 days of differentiation, the cells were treated with 100 ng/mL of LPS, and 24 h later, further transfected with different plasmids (20 ng/μL).
Silencing RNF7 plasmids (sh-RNF7) and overexpression RNF7 plasmids (oe-RNF7) were constructed, and the respective sets of empty vector plasmids were used as negative controls (NC) for overexpression (oe-NC, overexpression vector without any sequence inserted) or silencing (sh-NC, interference vector without any sequence inserted). All plasmids were constructed by RiboBio (Guangzhou, China). Cells were treated with LPS 100 ng/mL and transfected with plasmid at a concentration of 20 ng/μL after 24 h. Cells were divided into the control, LPS, LPS+sh-NC, LPS+sh-RNF7, LPS+oe-NC, LPS+oe-RNF7, and LPS+oe-RNF7+SC79 groups. The concentration of SC79 was 50 nM. The cells in each group were collected 48 h after transfection for subsequent assays.
Statistical analysis.
All data were processed using SPSS 21.0 statistical software (IBM Corp., Armonk, NY, USA). Measurement data were expressed as mean ± standard deviation. The data between two groups were analyzed by the independent sample t test. The data among multiple groups were analyzed by one-way analysis of variance with Tukey’s post hoc test. In all statistical references, a value of P < 0.05 was considered to be statistically significant.
Data availability.
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
Y.L. and X.J. designed the study. Y.L. and X.J. collated the data, carried out data analyses, and produced the initial draft of the manuscript. M.S. and Z.J. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Zhiyong Ji, Email: jizy@jlu.edu.cn.
Denise Monack, Stanford University.
REFERENCES
- 1.Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care Trialists . 2016. Assessment of global incidence and mortality of hospital-treated sepsis: current Estimates and Limitations Am J Respir Crit Care Med 193:259–272. 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 2.Perner A, Gordon AC, De Backer D, Dimopoulos G, Russell JA, Lipman J, Jensen JU, Myburgh J, Singer M, Bellomo R, Walsh T. 2016. Sepsis: frontiers in diagnosis, resuscitation and antibiotic therapy. Intensive Care Med 42:1958–1969. 10.1007/s00134-016-4577-z. [DOI] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, Vincent JL. 2016. Sepsis and septic shock. Nat Rev Dis Primers 2:16045. 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchese FP, Raimondi I, Huarte M. 2017. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18:206. 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C, Yang L. 2018. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol 28:287–301. 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Z, Li Y, Li Y, Pan J, Wang J, Fang F. 2020. Long noncoding RNA and messenger RNA abnormalities in pediatric sepsis: a preliminary study. BMC Med Genomics 13:36. 10.1186/s12920-020-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia Y, Li Z, Cai W, Xiao D, Han S, Han F, Bai X, Wang K, Liu Y, Li X, Guan H, Hu D. 2018. SIRT1 regulates inflammation response of macrophages in sepsis mediated by long noncoding RNA. Biochim Biophys Acta Mol Basis Dis 1864:784–792. 10.1016/j.bbadis.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang F, Liu X, Liu G, Qiu H, He Y, Hu H, Jiang P. 2020. Long non-coding RNA RNF7 promotes the cardiac fibrosis in rat model via miR-543/THBS1 axis and TGFβ1 activation. Aging (Albany NY) 12:996–1010. 10.18632/aging.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Audet GN, Fulks D, Stricker JC, Olfert IM. 2013. Chronic delivery of a thrombospondin-1 mimetic decreases skeletal muscle capillarity in mice. PLoS One 8:e55953. 10.1371/journal.pone.0055953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balboa E, Saavedra-Leiva F, Cea LA, Vargas AA, Ramirez V, Escamilla R, Saez JC, Regueira T. 2018. Sepsis-induced channelopathy in skeletal muscles is associated with expression of non-selective channels. Shock 49:221–228. 10.1097/SHK.0000000000000916. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Li TM, Liu XR, Bai YP, Li J, Tang N, Wang XB. 2019. MicroRNA-140 inhibits skeletal muscle glycolysis and atrophy in endotoxin-induced sepsis in mice via the WNT signaling pathway. Am J Physiol Cell Physiol 317:C189–C199. 10.1152/ajpcell.00419.2018. [DOI] [PubMed] [Google Scholar]
- 12.Stana F, Vujovic M, Mayaki D, Leduc-Gaudet JP, Leblanc P, Huck L, Hussain SNA. 2017. Differential regulation of the autophagy and proteasome pathways in skeletal muscles in sepsis. Crit Care Med 45:e971–e979. 10.1097/CCM.0000000000002520. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Liu H, Chu T, Jiang P, Li ST. 2019. Neuregulin-1beta attenuates sepsis-induced diaphragm atrophy by activating the PI3K/Akt signaling pathway. J Muscle Res Cell Motil 40:43–51. 10.1007/s10974-019-09512-2. [DOI] [PubMed] [Google Scholar]
- 14.Tan M, Xu J, Siddiqui J, Feng F, Sun Y. 2016. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer 15:81. 10.1186/s12943-016-0567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu S, Li H, Chen W, Zhao J, Gao L, Bo T. 2018. Beta-arrestin 1 mediates liver thyrotropin regulation of cholesterol conversion metabolism via the Akt-dependent pathway. Int J Endocrinol 2018:4371396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Cai J, Shi K, Li H, Du J, Hu D, Liu Z, Wang W. 2021. Germacrone induces lung cancer cell apoptosis and cell cycle arrest via the Akt/MDM2/p53 signaling pathway. Mol Med Rep 23:452. 10.3892/mmr.2021.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi B, Ma M, Zheng Y, Pan Y, Lin X. 2019. mTOR and Beclin1: two key autophagy-related molecules and their roles in myocardial ischemia/reperfusion injury. J Cell Physiol 234:12562–12568. 10.1002/jcp.28125. [DOI] [PubMed] [Google Scholar]
- 18.Loeffler DA. 2019. Influence of normal aging on brain autophagy: a complex scenario. Front Aging Neurosci 11:49. 10.3389/fnagi.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Froberg JE, Lee JT. 2014. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 39:35–43. 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JX, Xu X, Zhang S. 2019. Silence of long noncoding RNA NEAT1 exerts suppressive effects on immunity during sepsis by promoting microRNA-125-dependent MCEMP1 downregulation. IUBMB Life 71:956–968. [DOI] [PubMed] [Google Scholar]
- 21.Geng F, Liu W, Yu L. 2019. Potential role of circulating long noncoding RNA MALAT1 in predicting disease risk, severity, and patients' survival in sepsis. J Clin Lab Anal 33:e22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q, Yi X. 2018. Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt signaling pathway in rats with epilepsy. J Mol Neurosci 65:234–245. 10.1007/s12031-018-1093-3. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzo D, Duarte A, Mundinano J, Berguer P, Nepomnaschy I, Piazzon I. 2016. A B-cell superantigen induces the apoptosis of murine and human malignant B cells. PLoS One 11:e0162456. 10.1371/journal.pone.0162456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, Wu Z, He Y, Xiao Y, Xia C. 2020. Single and dual target inhibitors based on Bcl-2: promising anti-tumor agents for cancer therapy. Eur J Med Chem 201:112446. 10.1016/j.ejmech.2020.112446. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Ding Y, Ye N, Wild C, Chen H, Zhou J. 2016. Direct activation of Bax protein for cancer therapy. Med Res Rev 36:313–341. 10.1002/med.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Yang H, Li WJ, Liu YH. 2020. LncRNA MALAT1 promotes OGD-induced apoptosis of brain microvascular endothelial cells by sponging miR-126 to repress PI3K/Akt signaling pathway. Neurochem Res 45:2091–2099. 10.1007/s11064-020-03071-6. [DOI] [PubMed] [Google Scholar]
- 27.Gao T, Dai X, Jiang Y, He X, Yuan S, Zhao P. 2020. LncRNA HAND2-AS1 inhibits proliferation and promotes apoptosis of non-small cell lung cancer cells by inactivating PI3K/Akt pathway. Biosci Rep 40:BSR20201870. 10.1042/BSR20201870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Guo L, Zhao M, Zhang D, Xu H, Nie Q. 2019. The inhibition on MDFIC and PI3K/AKT pathway caused by miR-146b-3p triggers suppression of myoblast proliferation and differentiation and promotion of apoptosis Cells 8:656. 10.3390/cells8070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen Z, Hou W, Wu W, Zhao Y, Dong X, Bai X, Peng L, Song L. 2018. 6'-O-galloylpaeoniflorin attenuates cerebral ischemia reperfusion-induced neuroinflammation and oxidative stress via PI3K/Akt/Nrf2 activation. Oxid Med Cell Longev 2018:8678267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao Y, Wang L, Hu T, Yin D, He H, He M. 2021. Capsaicin protects cardiomyocytes against lipopolysaccharide-induced damage via 14-3-3γ-mediated autophagy augmentation. Front Pharmacol 12:659015. 10.3389/fphar.2021.659015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cicek M, Unsal V, Doganer A, Demir M. 2021. Investigation of oxidant/antioxidant and anti-inflammatory effects of apigenin on apoptosis in sepsis-induced rat lung. J Biochem Mol Toxicol 35:e22743. [DOI] [PubMed] [Google Scholar]
- 32.Chang SC, Ding JL. 2014. Ubiquitination by SAG regulates macrophage survival/death and immune response during infection. Cell Death Differ 21:1388–1398. 10.1038/cdd.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong X, Mathewson ND, Li H, Tan M, Fujiwara H, Li H, Reddy P, Sun Y. 2018. SAG/RBX2 E3 ubiquitin ligase differentially regulates inflammatory responses of myeloid cell subsets. Front Immunol 9:2882. 10.3389/fimmu.2018.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy JMM, Towers CG, Thorburn A. 2017. Targeting autophagy in cancer. Nat Rev Cancer 17:528–542. 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. 2018. Autophagy and inflammation. Annu Rev Immunol 36:73–101. 10.1146/annurev-immunol-042617-053253. [DOI] [PubMed] [Google Scholar]
- 36.Mofarrahi M, Sigala I, Guo Y, Godin R, Davis EC, Petrof B, Sandri M, Burelle Y, Hussain SN. 2012. Autophagy and skeletal muscles in sepsis. PLoS One 7:e47265. 10.1371/journal.pone.0047265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho J, Yu J, Wong SH, Zhang L, Liu X, Wong WT, Leung CC, Choi G, Wang MH, Gin T, Chan MT, Wu WK. 2016. Autophagy in sepsis: degradation into exhaustion? Autophagy 12:1073–1082. 10.1080/15548627.2016.1179410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu XY, Fang Y, Zheng FX, Zhang YZ, Li QL. 2020. LncRNA NEAT1 facilitates the progression of sepsis through up-regulating TSP-1 via sponging miR-370-3p. Eur Rev Med Pharmacol Sci 24:333–344. [DOI] [PubMed] [Google Scholar]
- 39.McMaken S, Exline MC, Mehta P, Piper M, Wang Y, Fischer SN, Newland CA, Schrader CA, Balser SR, Sarkar A, Baran CP, Marsh CB, Cook CH, Phillips GS, Ali NA. 2011. Thrombospondin-1 contributes to mortality in murine sepsis through effects on innate immunity. PLoS One 6:e19654. 10.1371/journal.pone.0019654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Morgan S, Ren J, Wang Q, Annis DS, Mosher DF, Zhang J, Sorenson CM, Sheibani N, Liu B. 2015. Thrombospondin-1 (TSP1) contributes to the development of vascular inflammation by regulating monocytic cell motility in mouse models of abdominal aortic aneurysm. Circ Res 117:129–141. 10.1161/CIRCRESAHA.117.305262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, Dong C, Chen X, Zhu R, Wang J. 2021. Silencing of miR-20b-5p exerts inhibitory effect on diabetic retinopathy via inactivation of THBS1 gene induced VEGF/Akt/PI3K Pathway. Diabetes Metab Syndr Obes 14:1183–1193. 10.2147/DMSO.S299143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Wang L, Meng L, Cao G, Wu Y. 2019. Sirtuin 6 overexpression relieves sepsis-induced acute kidney injury by promoting autophagy. Cell Cycle 18:425–436. 10.1080/15384101.2019.1568746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4 and Fig. S1 and S2. Download iai.00535-22-s0001.pdf, PDF file, 0.2 MB (220.8KB, pdf)
Data Availability Statement
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.