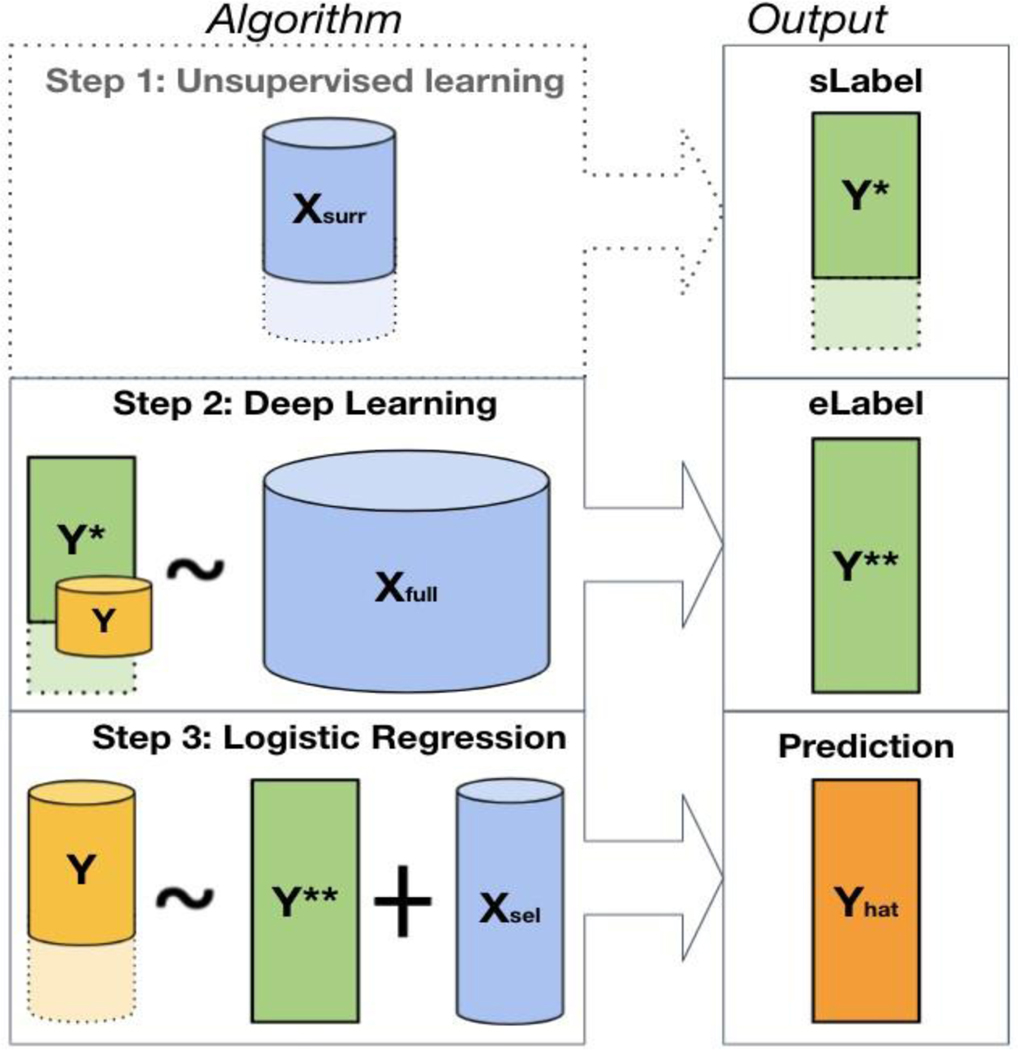
Figure 1.
The WSS-DL method pipeline. Step 1: creation of silver standard labels from using the MAP algorithm; this step may be omitted when is a viable surrogate that can be directly used as . Step 2: creation of the enhanced-silver-standard label using neural network, with as the outcome, as input feature, and a subset of for fine-tuning. Step 3: final prediction of using logistic regression, with the enhanced-silver-standard labels and a minimal set of informative features as input features. The different sizes of represented in steps 2 and 3 indicate that a mere subset of is included for algorithm fine-tuning in Step 2, and the full vector in Step 3 for the final patient-level phenotype classification.