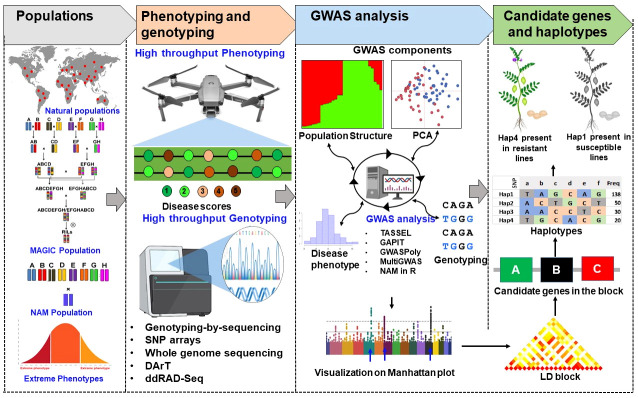
Figure 3.
Illustration of genome-wide associations studies to identify genes associated with disease resistance. The partially structured (NAM and MAGIC) and unstructured populations (germplasm lines, association panels) can be used for high throughput phenotyping and genotyping to perform high resolution association mapping with advance tools for genome wide association analysis (GWAS). The peaks identified in GWAS analysis can be used for identification of LD blocks. Each LD block includes one or few candidate genes associated with the trait can be used for validation or development of diagnostic markers for genomics associated breeding. The validated genes can be further used for identification of haplotypes for disease resistance or disease susceptibility.