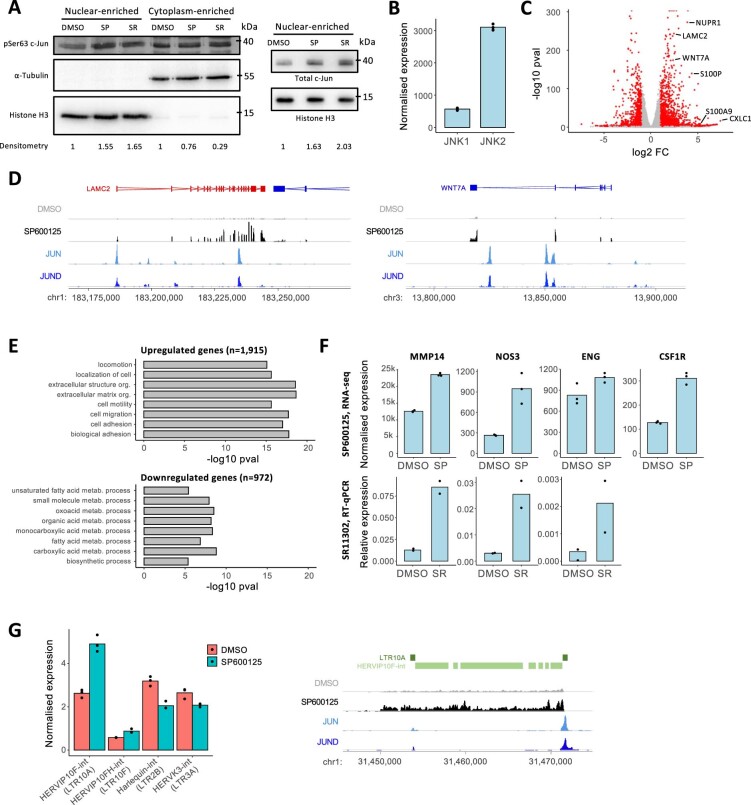
Extended Data Fig. 4. Effects of SP600125 treatment in hTSCs.
A, Western blot of phosphorylated c-Jun (pSer63 c-Jun) and total c-Jun in enriched nuclear or cytoplasmic fractions of cells treated with the AP-1 inhibitors SP600125 (‘SP’) or SR11302 (‘SR’). Densitometry values normalized to the respective loading controls are shown below. Full blot images are in Supplementary Figure 7. B, Expression of JNK1 (MAPK8) and JNK2 (MAPK9) in hTSCs (data from RNA-seq). C, Volcano plot of RNA-seq data from cells treated with SP600125. Some genes implicated in cell migration are highlighted. P values are from the DESeq2 R package. D, Genome browser snapshots showing examples of genes highlighted in C, together with CUT&Tag data for JUN and JUND. E Gene ontology biological processes enriched terms for genes up- or downregulated after treatment of hTSCs with SP600125. P values are from the topGO R package. F, Expression of MMP14 (a known AP-1 target), NOS3, ENG and CSF1R in hTSCs treated with SP600125 (data from RNA-seq) and/or SR11302 (see data from RT–qPCR). G, Expression of internal ERV fragments driven by LTRs of H3K27ac-enriched families (in brackets) upon SP600125 treatment. An example of LTR10A-driven transcription of a proviral HERVIP10-F locus is shown on the right. Please see the data availability statement for details on source data for panels B, C and E–G.