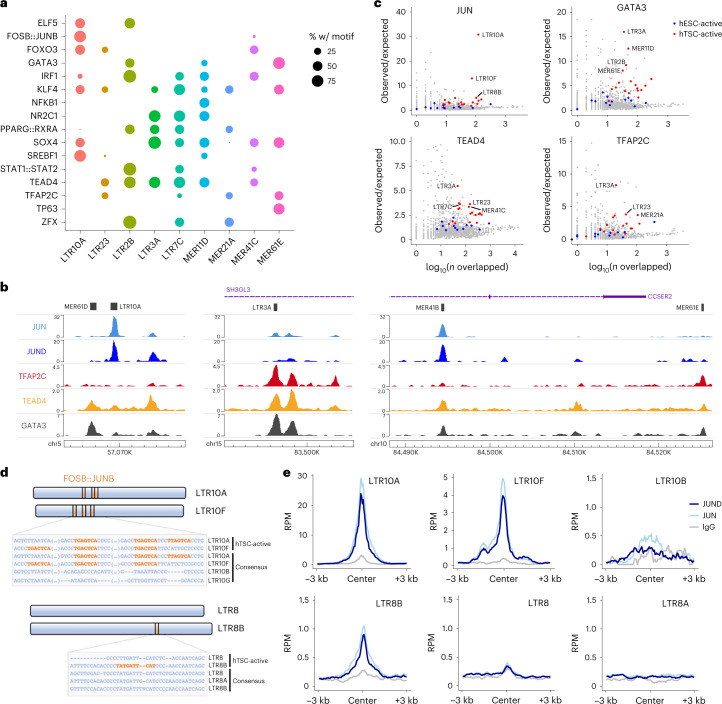
Fig. 2. Transcription-factor repertoire at hTSC-active ERVs.
a, Proportion of H3K27ac-marked elements from selected ERV families bearing motifs for the transcription factors on the y axis. b, Genome browser snapshots of transcription-factor CUT&Tag/RUN data, showing examples of enrichment over ERVs. c, Repeat family-wide enrichment for peaks from transcription-factor CUT&Tag/RUN data on hTSCs. H3K27ac-enriched families in hTSCs or hESCs are highlighted. d, Schematic and alignment of LTR10 and LTR8 subfamilies showing the presence of FOSB::JUNB (that is, AP-1) motifs in the genome-wide consensus sequence and/or in a consensus of the H3K27ac-marked elements. e, Mean CUT&Tag profiles for JUN and JUND over LTR10 and LTR8 subfamilies. See the data availability statement for details on source data for a, c and e.