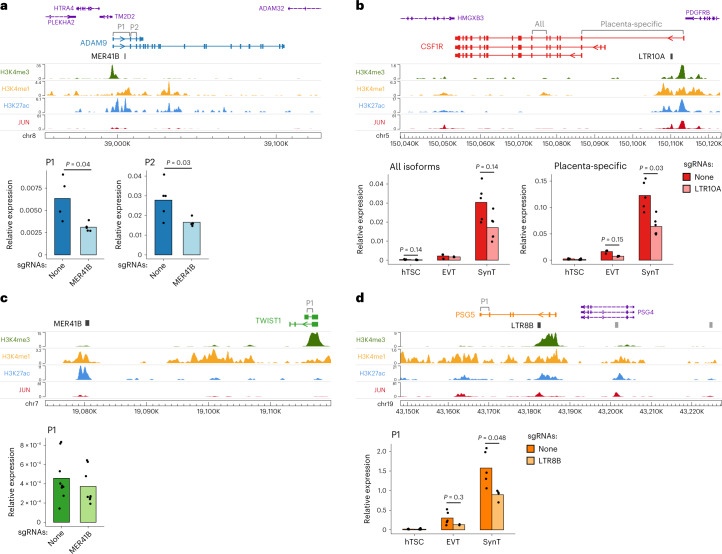
Fig. 5. Genetic excision of hTSC-active ERVs.
a, Genome browser snapshot of the ADAM9 locus containing an enhancer-like MER41B element, and ADAM9 expression (using primer pairs highlighted on the gene annotation) in hTSC populations treated with lentiviral CRISPR constructs carrying either no sgRNAs (n = 4) or sgRNAs that excise the MER41B element (n = 5 from two sgRNA sets). b, As in a, but for CSF1R and its intronic LTR10A elements. CSF1R expression was measured in hTSCs (n = 5 from one sgRNA set) and hTSC-derived EVT (n = 3) and SynT (n = 5 no sgRNA, n = 6 LTR10A sgRNAs). c, As in a, but for TWIST1 and a downstream MER41B element (n = 9 no sgRNA, n = 8 MER41B sgRNAs; from three infections using one sgRNA set). d, As in a, but for PSG5 and its intronic LTR8B elements. PSG5 expression was measured in hTSCs (n = 3 no sgRNA, n = 6 LTR8B sgRNAs from two sgRNA sets) and hTSC-derived EVT (n = 5 no sgRNA, n = 2 LTR8B sgRNAs) and SynT (n = 5 no sgRNA, n = 4 LTR8B sgRNAs). P values throughout are from two-sided Wilcoxon tests with multiple comparisons correction. See the data availability statement for details of the source data for RT–qPCR.