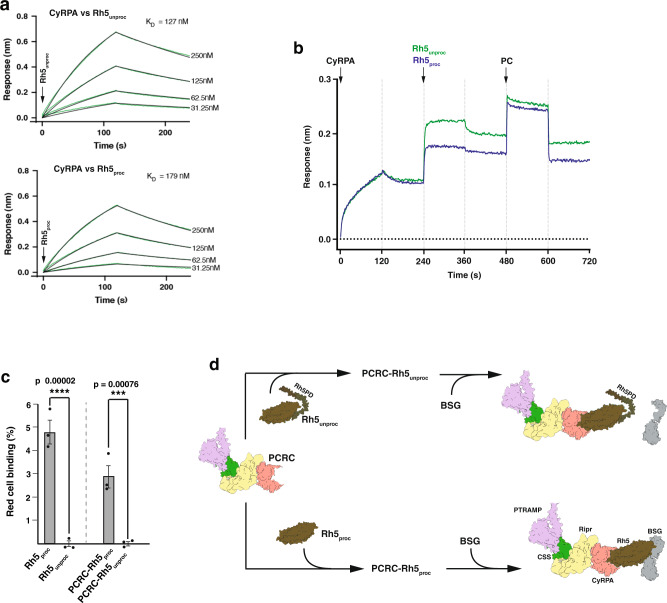
Fig. 5. The role of the N-terminal prodomain of PfRh5 in the PCRCR complex.
a Binding of CyRPA to either Rh5unproc or Rh5proc was determined by Biolayer Interferometry (BLI). Biotinylated CyRPA was pre-added to a Streptavidin biosensor, before dipping into twofold dilutions of Rh5unproc and Rh5proc at t = 0 s. Binding occurred for 120 s followed by dissociation for a further 120 s. The KD values were calculated using Octet Data Analysis v11.0 software. b Binding of Rh5unproc and Rh5proc to the RCR complex by BLI. CyRPA was added at t = 0 s to biotinylated Ripr immobilised on a Streptavidin biosensor for 120 s before allowing dissociation for a further 120 s. Either Rh5unproc or Rh5proc were added to Ripr-CyRPA and allowed to bind, then dissociate, before PTRAMP-CSS (PC) was added at t = 480 s. Binding and dissociation were allowed to occur until t = 720 s. c Binding of Rh5unproc or Rh5proc and PCRC-Rh5unproc or PCRC-Rh5proc to erythrocytes measured by flow-cytometric analysis. The assay was done three times and means (+/−) standard error of the mean (SEM) is shown. One-way ANOVA with Sidak’s multiple comparisons test was used to calculate p values. d Representation of the binding results in c using known structures of CSS (PDB ID: 7UNZ), PfRipr, CyRPA, PfRh5 (PDB ID: 6MPV), basigin (BSG) (PDB ID: 3B5H) and the AlphaFold Monomer v2.0 structures52 of the PTRAMP ectodomain (Uniprot: Q8I5M8) and Rh5PD (UniProt: Q8IFM5). Structures were assembled in ChimeraX (https://www.rbvi.ucsf.edu/chimerax). Source data are provided as a source data file.