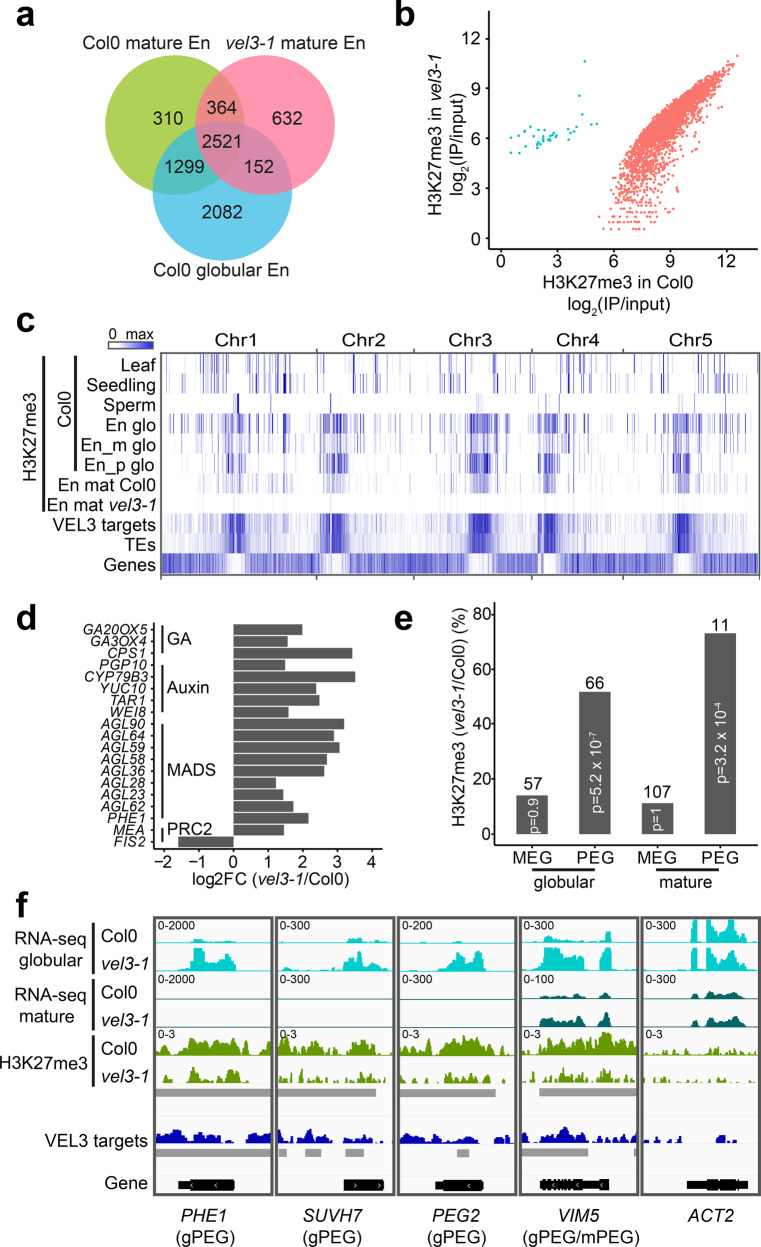
Fig. 4. VEL3 is required for the establishment of a persistent pattern of H3K27me3 deposition in the endosperm.
a Chromatin immunoprecipitation to compare the number of genes marked with H3K27me3 in mature Col-0 and vel3-1 endosperms, compared to globular stage endosperm tissues21, showing conservation of marks across endosperm development and dependency on VEL3. b Quantitative effect of VEL3 loss on H3K27me3 at genes in the mature endosperm shows a reduction in H3K27me3 genome wide at gene loci. Blue shows loci with increased H3K27me3 in vel3-1 vs. Col-0, red shows loci with reduced H3K27me3 in vel3-1. c Genome-wide comparison of VEL3-GFP chromatin distribution with H3K27me3, location of genes and transposons (TEs), demonstrating the general requirement for VEL3 for the targeting of H3K27me3 to pericentromeric regions bound by the VEL3 protein. En glo: globular endosperm; En_m glo: maternally derived endosperm alleles at globular stage; En_p paternally derived alleles; TE: transposable element; En mat: mature endosperm. d RNA-seq analysis of WT vs. vel3-1 mutant seeds at the globular stage at 16 °C. Expression of genes known to be mis-regulated in endosperms lacking FIS2-PRC2 are shown, demonstrating a role for VEL3 in regulating genes silenced by PRC2. All genes shown are significantly mis-expressed in vel3-1 compared to WT (FDR < 0.05) in three biological replicate experiments. e VEL3 is required for H3K27me3 deposition and the establishment of PEGs in the endosperm. Chart shows the percentage of PEGs and MEGs at globular stage and mature stage endosperms that show VEL3-dependent H3K27me3 deposition. The number of genes in each category is shown above the bars. Significantly enriched loci were determined using two-sided Fisher’s exact test. f VEL3 is required for normal expression levels of PEGs. RNA-seq comparing WT and vel3-1 mutants shown with H3K27me3 levels and VEL3 protein binding sites at four previously characterized PEGs and the control gene ACTIN2 (ACT2). gPEGs are PEGs at the globular stage, mPEGs are PEGs in mature seeds. Gray bars indicate significant regions (p < 0.05) identified by DiffBind (for H3K27me3 between Col0 and vel3-1) or MACS2 (for VEL3 ChIP between IP and input).