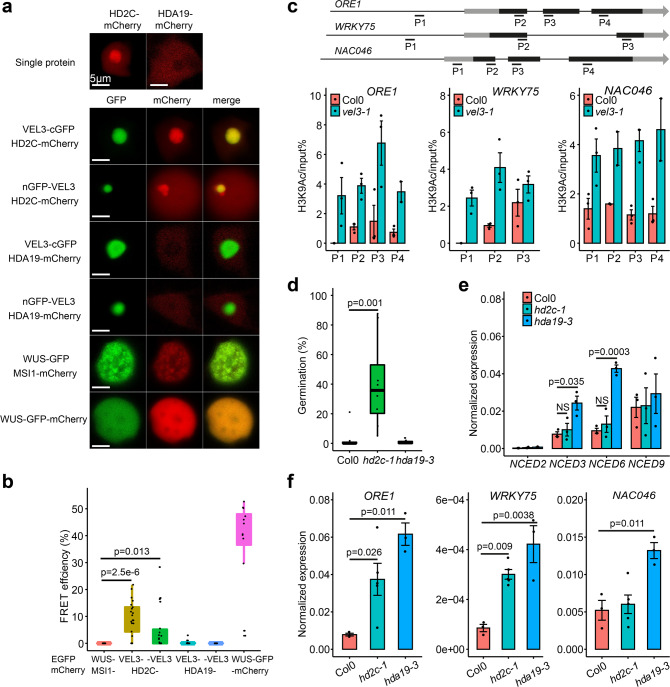
Fig. 6. VEL3 is required for the function of histone deacetylase complexes in the endosperm regulating seed dormancy.
a VEL3 co-localizes with HD2C in the nucleolus, but not with HDA19, which exists pre-dominantly in the nucleoplasm during transient expression in Nicotiana. b FRET assays show that VEL3 and HD2C associate in planta. VEL3-: VEL3 C-terminal GFP fusion. -VEL3: VEL3 N-terminal GFP fusion. Self-dimerizing WUS-GFP/mCHERRY is shown as the positive control. Boxplots indicate minimum and maximum values, as well as 25th, 50th, and 75th quartiles. p-values are derived from the two-sided t-test. Sample sizes are included in source data (N = 16,21,18,16,16,12 from left to right). The experiments were repeated twice. c VEL3 is required for H3K9 deacetylation at ORE1, WRKY75 and NAC046. Gene models show location of exons in black and primers used for ChIP. Charts show H3K9Ac levels in mature endosperms of seeds set at 16 °C. Data shown as mean +/– SE of three biological replicate immunoprecipitations tested by qPCR. d Loss of HD2C causes a similar effect on seed dormancy to loss of VEL3 in seeds set at 16 °C, but hda19-3 mutants show a WT phenotype. Data represents mean +/– SE of 8 to 12 biological replicate seed batches per genotype. Boxplots indicate minimum and maximum values, as well as 25th, 50th, and 75th quartiles. p-values are derived from the two-sided t-test. e Analysis of NCED gene expression in WT, hd2c-1 and hda19-3 mutants set at 16 °C. Data shown as mean +/– SE of three biological replicates tested by qRT-PCR. p-values are derived from the two-sided t-test. f qRT-PCR analysis of the expression of ORE1, WRKY75 and NAC046 in hd2c-1 and hda19-3 mutants. Data shown as mean +/– SE of three biological replicates. p-values are derived from the two-sided t-test.