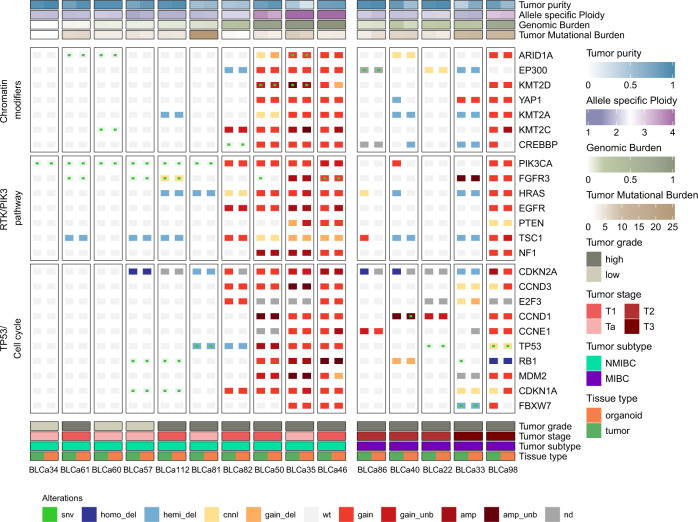
Fig. 4. Tumor and matched patient-derived organoids (PDOs) recapitulate typical mutational mechanisms of bladder cancer (BLCa).
Mutation heat-map. Samples are represented in the columns, with primary tumor on the left (green; n = 15 biological samples) and PDOs on the right (orange; n = 15 biological samples). PDO/PT pairs from the same samples are grouped by tumor subtype and ordered by increasing genomic burden within groups. The copy-number profile of the samples with high allele-specific Ploidy (asP, see “Methods”) is characterized by universal copy-number gains and amplifications. Rows represent genes grouped per pathway. Different types of genomic alterations are indicated in different colors in the bottom and Tumor purity, allele-specific Ploidy, Genomic Burden, and Tumor Mutational Burden are reported on the top. amp amplification, amp_unb amplification unbalanced, cnnl copy number neutral loss, gain_del gain deletion (gain with Loss of heterozygosity), gain_unb gain unbalanced, homo_del homo-deletion, hemi_del hemi-deletion, MIBC muscle-invasive BLCa, NMIBC non-muscle invasive BLCa, nd not determined, snv single nucleotide variant, wt wild-type.