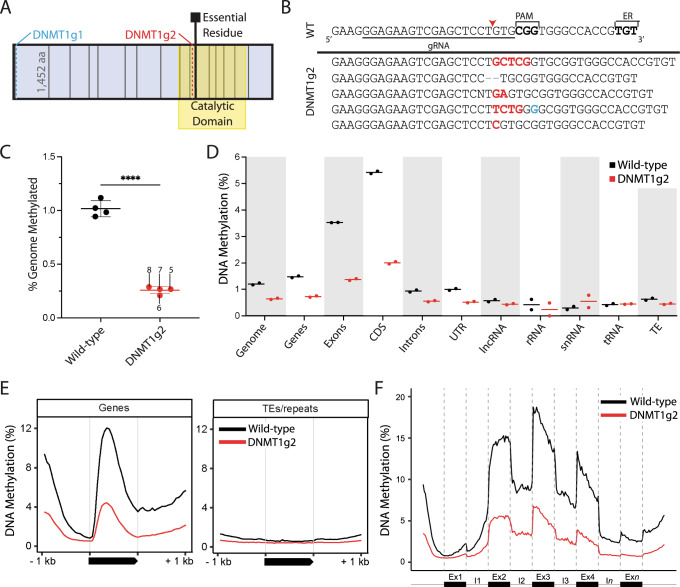
Fig. 1. Mutations in the DNMT1 catalytic domain result in decreased DNA methylation.
A Clonal raider ant DNMT1 protein model. Vertical solid lines indicate exon boundaries; broken lines indicate CRISPR/Cas9 target cut sites. The DNMT1g1 (blue) target site is early in the second exon, while the DNMT1g2 (red) target site is in the catalytic domain (yellow), upstream of an essential cysteine residue (black square). B Top: Wild-type (WT) DNMT1 sequence at the DNMT1g2 target site, with the codon for the essential residue (ER) and the protospacer adjacent motif (PAM) in bold. Guide-RNA (gRNA) sequence underlined. Red arrowhead indicates predicted cut site. Bottom: Mutant alleles observed in G0 adults show insertions (red), deletions (gray) and base changes (blue). C DNMT1g2 mutants have decreased genome-wide methylation compared to wild-type ants reared in parallel, consistent with a functional defect in DNMT1 (two sided unpaired T-test: p < 0.0001; n = 4 animals per condition). Error bars show standard deviation around the mean. Each data point represents low coverage WGBS of DNA extracted from the remaining tissue of a single ant after brain and ovary dissection. Data point labels correspond to animals #5-8 in Supplementary Table 1. Methylation levels are corrected for bisulfite non-conversion. D Percentages of methylated cytosines for different genomic features based on high coverage WGBS of two biological replicates of wild types (black) and DNMT1g2 mutants (red) from (C) (bars denote means). E DNA methylation patterns in wild types (black) and DNMT1g2 mutants (red) for genes and TEs/repeats. The 1 kb upstream and downstream regions along with the gene body region (black bar) are displayed. F Gene body methylation profile for different positions of exons and introns across the O. biroi genome. Exons and introns are shown separately for wild types (black) and DNMT1g2 mutants (red). Source data are provided as a Source Data file.