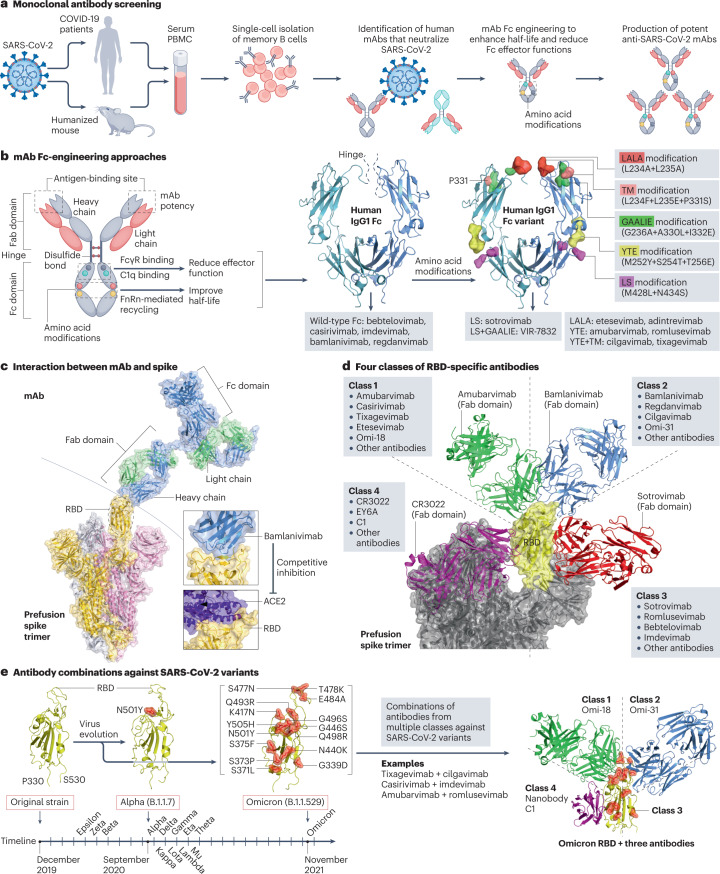
Fig. 3. Development of anti-spike monoclonal antibodies.
a, Screening of monoclonal antibodies (mAbs). Peripheral blood mononuclear cells (PBMCs) are collected from convalescent donors or humanized mice exposed to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Single-cell cultures of plasma or memory B cells are prepared to screen potent mAbs. b, Fragment crystallizable (Fc)-engineering approaches that modify amino acids in the Fc domain of mAbs to enhance the half-life and reduce the effector functions38. Common modifications such as LALA, TM, LS, YTE and GAALIE are indicated, with their wild-type residues visualized using the structure of human IgG1 Fc (PDB: 4X4M), and their applications in selected mAbs are shown below. c, Structure of a mAb in complex with the prefusion spike trimer. Bamlanivimab (PDB: 7KMG) blocks the binding of angiotensin-converting enzyme 2 (ACE2) to the receptor-binding domain (RBD) of spike (PDB: 6M17). d, Four antibody classes can be defined based on their interactions with the spike RBD. Representative antibodies from each class are shown, including amubarvimab (class 1, PDB: 7CDI), bamlanivimab (class 2, PDB: 7KMG), S309 (class 3, PDB: 7TNO) and CR3022 (class 4, PDB: 6W41). e, Antibodies from multiple classes can be combined to inhibit SARS-CoV-2 variants. SARS-CoV-2 variants harbour amino acid substitutions in the RBD (for example, 15 mutations in the Omicron variant) that potentially hamper antibody potency. The timeline of the earliest documented samples provided by the WHO (see Related links) shows viral evolution from the SARS-CoV-2 original strain to more than ten variants to date, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Epsilon (B.1.427/B.1.429), Zeta (P.2), Eta (B.1.525), Theta (P.3), Iota (B.1.526), Kappa (B.1.617.1), Lambda (C.37), Mu (B.1.621) and Omicron (B.1.1.529). A three-antibody combination of Omi-18 from class 1, Omi-31 from class 2 and nanobody C1 from class 4 can simultaneously target one RBD of an Omicron variant (PDB: 7ZFB). Fab, fragment antigen binding.