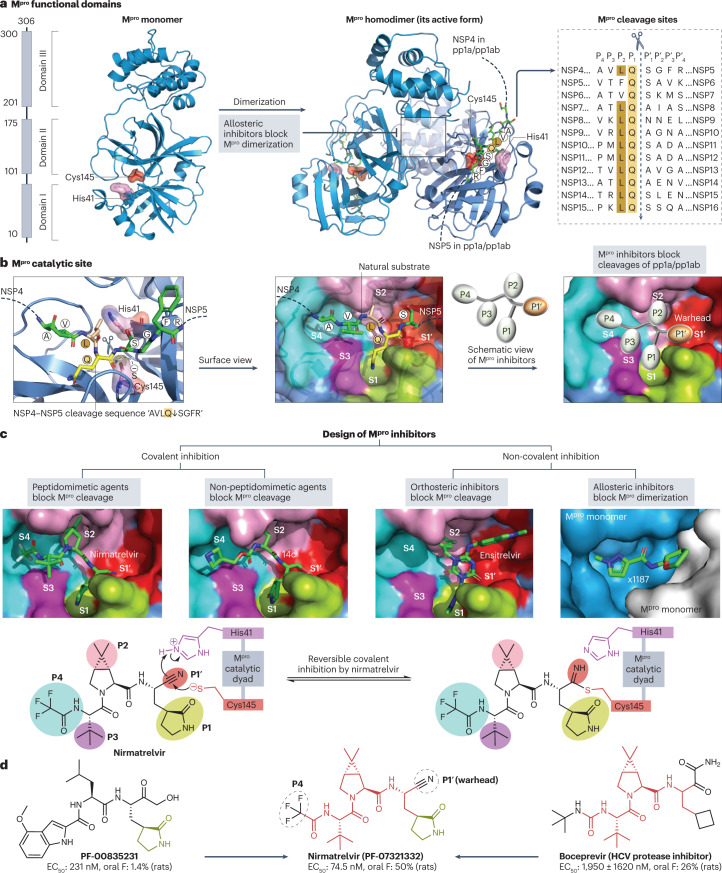
Fig. 4. Structure of the SARS-CoV-2 main protease and its drug-binding pocket.
a, The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (Mpro) functional domains, protein structures and cleavage sequences. The Mpro homodimer is the active form (PDB: 7VU6). The cleavage sequence ‘AVLQ↓SGFR’ between adjacent NSP4 and NSP5 in the pp1a and pp1ab polyproteins is localized across the Mpro catalytic dyad formed by Cys145 and His41 (PDB: 7DVP). Mpro cleavage sequences from the reference genome are shown on the right. b, Mpro catalytic site in the pre-cleavage state with the NSP4−NSP5 cleavage sequence (PDB: 7DVP). Mpro inhibitors with P1′ warhead, P1, P2, P3 and P4 moieties can be developed to maximize the drug–receptor interactions at the S1′, S1, S2, S3 and S4 subsites of Mpro, respectively. c, Four classes of Mpro inhibitors and the drug-binding pockets of nirmatrelvir (PDB: 7VH8), 14c (PDB: 7T4B), ensitrelvir (PDB: 7VU6) and x1187 (PDB: 5RFA). Reversible covalent inhibition of nirmatrelvir via the catalytic dyad Cys145−His41 is also shown77. The drug-binding pocket of x1187 is captured at the dimer interface (see panel a). d, Development of nirmatrelvir from PF-00835231 (ref. 78). Nirmatrelvir and boceprevir share identical structures at the backbone and P2/P3 moieties. The half-maximal effective concentration (EC50) values of PF-00835231, nirmatrelvir and boceprevir against SARS-CoV-2 USA_WA1/2020 and oral bioavailability (oral F) in rats were obtained from the literature78,282.