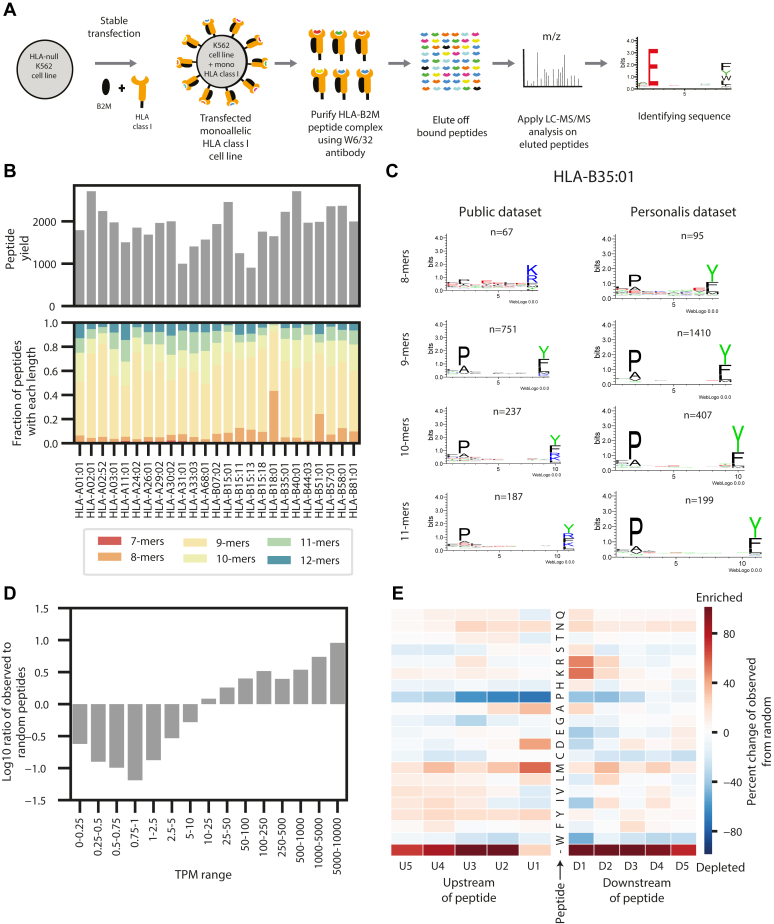
Fig. 1.
Generation and overview of the monoallelic data.A, a schematic of the experimental procedure to generate the monoallelic training data. A human leukocyte antigen (HLA) allele and beta 2 microglobulin (B2M) were stably transfected into an HLA-null K562 parental cell line. The major histocompatibility complex (MHC)–peptide complex was purified using a w6/32 antibody, and the peptides were gently eluted off the complexes. The peptides were sequenced with LC–MS/MS and identified with a database search. B, bar plots showing the peptide yields and distribution of peptide lengths for each of the 25 monoallelic cell lines. C, a comparison between motifs of peptides generated from our monoallelic cell line with HLA-B35:01 and a publicly available dataset for the same allele. Motifs are shown for peptides of length 8, 9, 10, and 11. See supplemental Fig. S1 for motifs from all 25 cells. See supplemental Fig. S2 for comparisons with other public datasets. D, a bar plot showing the distribution of the ratio of observed peptides from the monoallelic cell lines compared with random expectation across several transcript per million (TPM) ranges. Values are shown with a log10 transformation. E, a heatmap showing the enrichment and depletion of five amino acids upstream and downstream of the peptides identified from the monoallelic cell lines compared with a random expectation. Red denotes the enrichment of amino acids, and blue denotes the depletion of them. The C and N terminus of the protein are denoted with “-.”