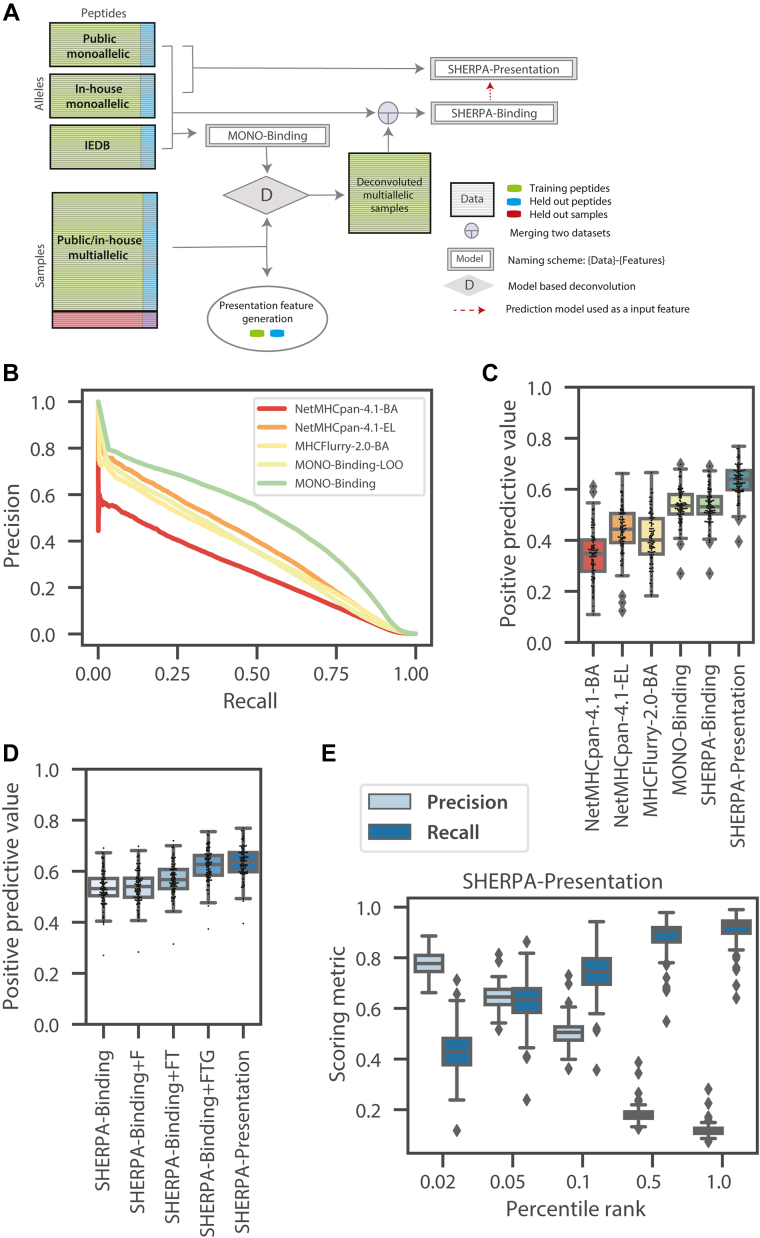
Fig. 5.
Overview of composite modeling approach and model performance.A, a schematic of the composite modeling approach. Inhouse monoallelic immunopeptidomics data, public monoallelic immunopeptidomics data, and Immune Epitope Database (IEDB) data are used to train MONO-Binding. MONO-Binding is used to deconvolute the multiallelic immunopeptidomics data to create pseudo monoallelic data. All monoallelic and pseudomonoallelic data are combined to train the Systematic Human Leukocyte Antigen Epitope Ranking Pan Algorithm (SHERPA) (SHERPA)-Binding model. The SHERPA-Binding model is used as a feature along with other presentation features to train the SHERPA-Presentation model on monoallelic immunopeptidomics data. B, a precision-recall curve demonstrating the predicted pan-performance on unseen alleles (MONO-Binding-LOO) compared with MONO-Binding and NetMHCpan4.1-BA, NetMHCpan-4.1-EL, MHCFlurry-2.0-BA. A model was trained for each allele with the data for that allele excluded from the training dataset. The MONO-Binding-LOO curve represents the predictions from each of the models on the test data of the allele excluded from the training data. C and D, boxplots denoting the distributions of positive predictive values (top 0.1%) across alleles within the monoallelic immunopeptidomics held-out test data. Distributions are shown for (C) NetMHCpan4.1-BA, NetMHCpan-4.1-EL, MHCFlurry-2.0-BA, MONO-Binding, SHERPA-Binding, and SHERPA-Presentation and (D) SHERPA-Binding, SHERPA-Binding + F, SHERPA-Binding + FT, SHERPA-Binding + TTG, and SHERPA-Presentation. E, boxplots showing the distribution of precision and recall values across alleles in the monoallelic immunopeptidomics data for SHERPA-Presentation across several percentile rank thresholds. A percentile rank of 0.1 is selected as the optimal threshold.