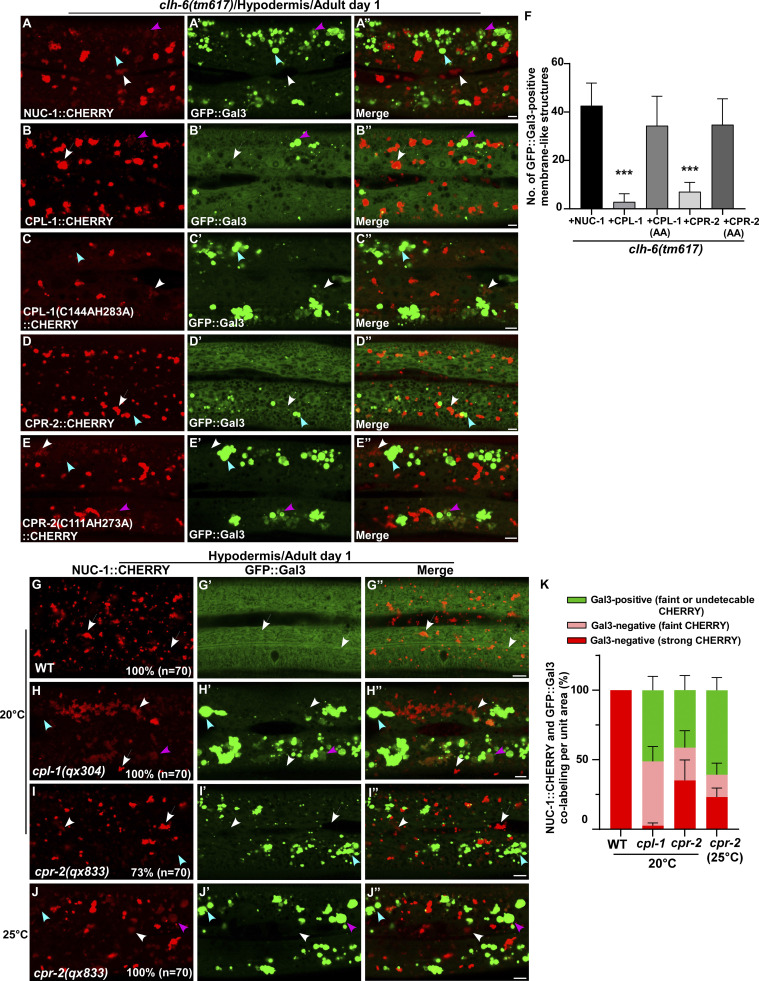
Figure 6.
CPL-1 and CPR-2 overexpression suppresses accumulation of damaged lysosomes in clh-6 mutants. (A–E′′) Confocal fluorescence images of the hypodermis in clh-6(tm617) co-expressing GFP::Gal3 and NUC-1::CHERRY (A–A′′), wild-type CPL-1::CHERRY (B–B′′), catalytically inactive CPL-1::CHERRY (C–C′′), wild-type CPR-2::CHERRY (D–D′′), or catalytically inactive CPR-2::CHERRY (E–E′′). CPL-1::CHERRY and CPR-2::CHERRY exhibit a vesicular pattern like NUC-1::CHERRY. (G–J′′) Confocal fluorescence images of the hypodermis in the indicated strains co-expressing NUC-1::CHERRY and GFP::Gal3. In A–E′′ and G–J′′, intact lysosomes contain strong NUC-1::CHERRY fluorescence and are not labeled by GFP::Gal3 (white arrows), while damaged lysosomes have either faint NUC-1::CHERRY fluorescence with GFP::Gal3 labeling (purple arrowheads) or without GFP::Gal3 labeling (white arrowheads), or strong GFP::Gal3 fluorescence with undetectable NUC-1::CHERRY (blue arrowheads). (F and K) Quantification analyses are shown in F and K. At least 10 animals were scored in each strain and data are shown as mean ± SD. In F, unpaired two-tailed Student’s t test was performed to compare datasets from CPL-1– or CPR-2–expressing worms with the NUC-1–expressing strain. ***P < 0.0001. All other points had P > 0.5. Scale bars: 5 µm.