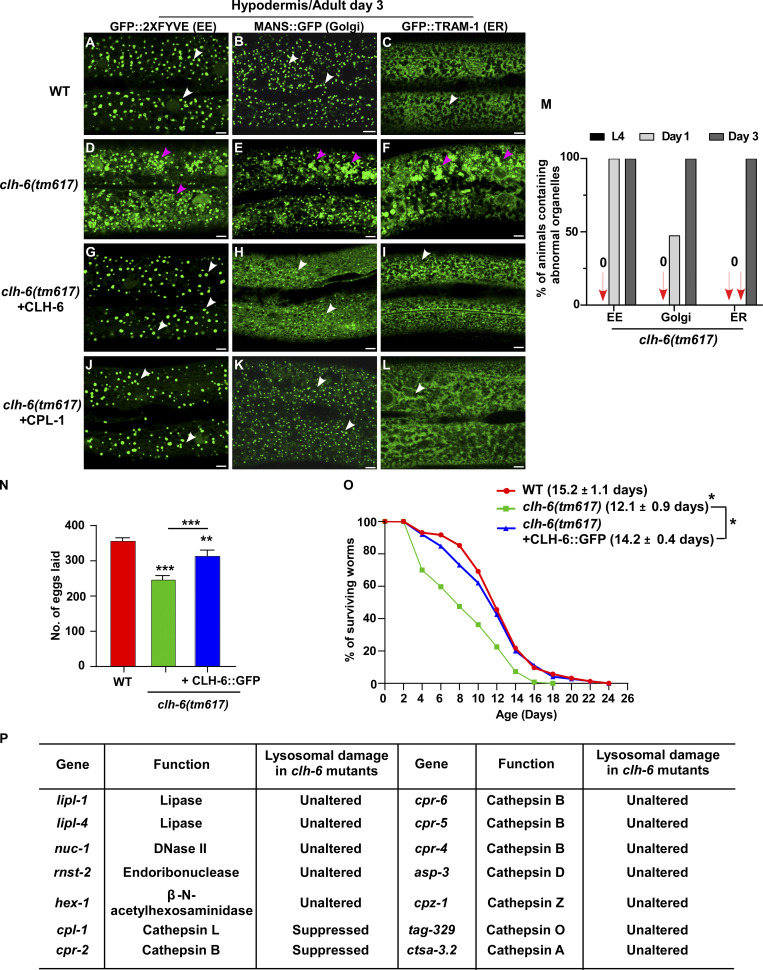
Figure S3.
Loss of clh-6 affects endosome, ER and Golgi patterns. (A–L) Confocal fluorescence images of the hypodermis in the indicated strains expressing different organellar markers. The punctate pattern of early endosomes (EE) and Golgi and the network pattern of ER (white arrows) are disrupted in clh-6(tm617) mutants (purple arrowheads) and restored by overexpression of CLH-6 and CPL-1. (M) The abnormal pattern of early endosomes, Golgi, and ER was quantified in clh-6(tm617) mutants at larval and adult stages. At least 30 animals were scored for each organellar marker in each stage. (N and O) Brood size and lifespan analyses were performed in the indicated strains. >15 and >100 worms were quantified in brood size and lifespan analyses, respectively. At least three independent experiments were performed, and data are shown as mean ± SD. Unpaired two-tailed Student’s t test was performed to compare mutant datasets with wild type or datasets that are linked by lines. *P < 0.05, **P < 0.001, ***P < 0.0001. Scale bars: 5 µm. (P) Lysosomal hydrolases that were tested by overexpression in clh-6(tm617) mutants. Lysosomal damage was examined by comparing accumulation of Gal3-positive structures in clh-6(tm617) mutants without and with the expression of each lysosomal hydrolase.